Professional Documents
Culture Documents
Marker-Assisted Foreground Selection For Identification of Salt Tolerant Rice Genotypes
Uploaded by
Jubair Al-rashidOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Marker-Assisted Foreground Selection For Identification of Salt Tolerant Rice Genotypes
Uploaded by
Jubair Al-rashidCopyright:
Available Formats
The Agriculturists 10(2): 1-8 (2012) A Scientific Journal of Krishi Foundation
ISSN-1729-5211 Indexed Journal
Marker-assisted Foreground Selection for Identification of Salt Tolerant Rice Genotypes
M. S. Alam 1*, M. Salim2 , M. Moniruzzaman3 , J. A. Rashid4 and M. M. Islam5
Regional Agricultural Research Station, BARI , Akbarpur, Moulvibazar, Bangladesh 2 Hill Agricultural Research Station, BARI , Khagrachari, Bangladesh 3 Bangladesh Jute Research Institute (BJRI), Manikgonj, Dhaka, Bangladesh 4 MATEX Bangladesh Ltd. Dhaka, Bangladesh 5 Biotechnology Div., Bangladesh Institute of Nuclear Agriculture, Mymensingh, Bangladesh *Corresponding author and Email: asarowar04bau@ gmail.com Received: 20 May 2012 Abstract Marker Assisted Selection (MAS) technique was used to develop salt tolerant rice genotypes using molecular markers during June 2009 to November 2010 in the experimental field and Biotechnology Laboratory of Plant Breeding Division, Bangladesh Institute of Nuclear Agriculture (BINA), Mymensingh. FL-378 was identified as donor or male parent for saltol QTL and Binadhan-7 as recurrent or recipient parent which had high yield with short life cycle. Crossing was done between them and 10 F1 seeds were produced. PCR bands from all the 10 F1 plants were scored as H represented heterozygous alleles for donor and recipient parent. Backcrossing was done to produce 105 BC1F1 seedlings. Foreground selection was performed with 72 BC1F1 plants with a tightly linked salt tolerance marker RM21. Out of 72 plants, 33 plants were selected for RM21 showing H scores. The selected segregants were subjected to further recombinant and background selections at BC1F1 generation. These selected genotypess could be used for further foreground, recombinant and background selections with appropriate markers upto BC3 generation for the development of salt tolerant rice genotypes. Keywords: Marker-Assisted Selection (MAS), foreground selection, salinity, SSR marker 1. Introduction Salinity is one of the major obstacles to increasing production in rice growing areas worldwide. Therefore, development of salt tolerant varieties has been considered as one of the strategies to increase rice production in salinity prone coastal areas. Salt stress is a major constraint to cereal production worldwide. In Bangladesh, out of 2.8 million hectares of the coastal area, around 1.0 million ha has become saline due to heavy withdrawal of surface and groundwater for irrigation and intrusion of seawater. The total coastal saline area covers one third of the 9 million hectares of total cultivated area in Bangladesh (ABSPII, 2006). Agriculture is a major sector of Bangladesh economy and the coastal area of Bangladesh is very fertile for growing rice. Increase in salinity intrusion and increase in soil salinity will have serious negative impacts on agriculture. The food production does not seem to have a better future in the event of a climate change. In Bangladesh, rice production may fall by 10 % and wheat by 30 % by 2050 (IPCC, 2007). Accepted: 15 November 2012
1
2 SSR or microsatellite markers are proved to be ideal for making genetic maps (Islam, 2004; Niones, 2004), assisting selection (Bhuiyan, 2005) and studying genetic diversity in crop germplasm. Microsatellite marker analysis is promising to identify major gene locus for salt tolerance that can be helpful for plant breeders to develop new cultivars. Currently, SSRs are predominantly being used to map and introgress agronomically important Quantitative Trait Loci (QTLs) into popular varieties using MarkerAssisted Backcrossing (MABC). However, their use is still limited due to lack of sufficient polymorphism particularly within related genotypes, labor requirements and cost of application. In addition, SSR markers have a low potential for multiplexing and require lengthy periods of time to genotype many markers as during the initial background selection in the marker-assisted backcrossing protocol (Thomson et al., 2010). Thus, the objective of this study was to identify salt tolerant rice genotypes using molecular markers and to introgress saltol gene into Binadhan-7 with marker assisted selection technique. 2. Materials and Methods 2.1. Field experiment Salt tolerant genotype FL-378 was selected as donor parent and Binadhan-7 was selected as recurrent or recipient parent. The experiment was conducted on the plastic pot (10L bucket) and pans designed by IRRI. The plants were labeled with pot labeler. 2.1.1. Location and time For the identification of salt tolerant rice genotypes with marker assisted backcrossing (MABC) technique, the experiment was conducted during the period from July 2009 to November 2010 in the experimental field and Biotechnology Laboratory of Plant Breeding Division, Bangladesh Institute of Nuclear Agriculture (BINA), Mymensingh.
Alam et al. /The Agriculturists 10(2): 1-8 (2012) 2.1.2. Plant materials Binadhan-7 was an early and high yielding salt susceptible mutant variety which was crossed with FL 378 which was an advanced salt tolerant line from IRRI. Binadhan-7 was the recurrent parent and FL 378 was the non-recurrent donor line. Seeds of selected F1 lines were grown in the pot. Then F1 plants were crossed with Binadhan7 as a principle of backcrossing and 150 BC1F1 seeds were developed. Among these seeds, 105 seeds were germinated in the experimental field in the next season. Among these 105 seedlings 72 seedlings were selected to collect the leaf samples to apply foreground selection using SSR markers. 2.1.3. F1 plant production Hybridization was carried out between Binadhan-7 and FL-378 . Binadhan-7 was used as female parent while FL-378 as male parent in the hybridization scheme. Three sets parental lines were seeded at an interval of 14 days starting from July 5, 2009. Twenty- one days old seedlings were transplanted in the hybridization block. a. Cultural nursery management of hybridization
Complete N-P-K-S fertilizers were applied at recommended doses. Urea were applied both at 20 and 40 DAT. Weeding and rouging were done carefully. Other cultural management practices were carried out as required. 2.1.4. Production of backcross seeds a. BC1 F1 seeds Production For producing BC1F1 seeds, F1 seeds were seeded on 15 February, 2010. Four sets of Binadhan-7 were seeded for synchronization of flowering with F1 plants for producing BC1F1 seeds. They were sown at an interval of 7 days starting from February 1, 2010.
Marker-assisted selection for salt tolerant rice b. Soaking, seeding and transplanting of BC1F1 seeds The total number of BC1F1 seeds produced was 150. In order to handle all the selection approaches and also for introgression of saltol gene by backcrossing all the BC1F1 seeds were used. 2.1.5. Raising of backcross plants The mature backcrossed seeds were collected 21 days after dusting. The seeds were dried in the oven at 500 C temperature for 72 hours for breaking dormancy. Seeds were seeded and then transplanted 21 days after seeding in the prepared pots (Fig. 2). 2.1.6. Collection of leaf sample for DNA extraction Young vigorously growing fresh leaf samples from these seedlings were collected from 25 day old seedlings to extract genomic DNA. Initially, healthy portion of the youngest leaves of the tiller were cut apart with sterilized scissors and washed in distilled water and ethanol (70%) and dried on fresh tissue paper to remove spore of microorganisms and any other source of foreign DNA. The collected leaf samples were then kept in polythene bags, avoiding any damage of the leaf tissues the bags were placed in an ice box to carry. Finally, the samples were stored in -800 C freezer with marking. 2.2. Genomic DNA extraction DNA was extracted from the leaves of each genotype using the Cetyl Trimethyl Ammonium Bromide (CTAB) mini-prep method at Biotechnology Lab., Bangladesh Institute of Nuclear Agriculture (BINA), Mymensingh. The simplified mini scale procedure for DNA isolation in PCR analysis developed at IRRI was followed. The quality of the isolated DNA in the protocol was sufficient for PCR analysis (Zheng et al., 1995). The following steps were followed in PCR-based DNA marker analysis.
3 2.3. PCR analysis for microsatellite (SSR) markers 2.3.1. Polymorphism survey for selection primer
Polymorphism survey of parent plants were carried out using 30 microsatellite markers. Out of these SSR primers, one primer (RM21) showed clear polymorphisms which were used in genotyping for the foreground selection of the 72 BC1F1 plants. The details of the primer are given in Table 1. 2.3.2. Polymerase chain reaction (PCR) The PCR cocktail including DNA had total volume of 14.75 l/reaction based on rice protocol, was placed in the PCR tubes and run in the DNA thermal cycler. The PCR reactions were: initial denaturation at 94C for 5 minutes, then final denaturation at 94C for 1 minute and annealing at 55C for one minute. Polymerization was carried out at 72C for 2 minutes to complete a cycle and cycle was repeated for 34 times. The final extension was at 72C for 7 minutes. After PCR, products were mixed with 3 l of 2X gel loading dye. Polymorphisms in the PCR products were detected by ethidium bromide staining after electrophoresis on 1.5% agarose gel using UV transilluminator. 2.3.3. Components of PCR cocktail (for 74 reactions) for foreground selection The following components were used to prepare PCR cocktail (Table 2). The total volume of PCR cocktail for this study was 12.75 l per sample. 2.4. Selection strategy for marker assisted selection (MAS) The use of DNA markers in backcrossing greatly increases the efficiency of selection. Three general levels of marker-assisted backcrossing (MABC) were described (Holland 2004). In the first level, markers can be used in combination
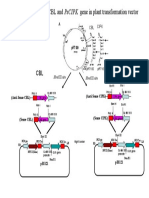
4 with or to replace screening for the target gene or QTL. This is referred to as foreground selection (Hospital and Charcosset, 1997). This may be particularly useful for traits that have laborious or time-consuming phenotypic screening procedures. It can also be used to select for reproductive-stage traits in the seedling stage, allowing the best plants to be identified for backcrossing. Furthermore, recessive alleles can be selected, which is difficult to do using conventional methods (Fig. 1). 2.5. Band scoring of F1 plants DNA samples were collected from 10 F1 plants and PCR was carried out using one SSR marker RM21. PCR bands from all the 10 F1 plants were scored as H. Score H represented heterozygous alleles for donor and recipient parent (Fig. 3). 2.6. Allele scoring for foreground selection The size (in nucleotide base pairs) of the most intensely amplified band for each microsatellite marker was determined based on its migration relative to a molecular weight size marker (1kb+ DNA ladder) with the help of Alpha Ease 4.0 software. The band having same level of Binadhan-7 was scored as A which indicated the homozygous allele of the recipient parent for the particular SSR marker. Again, the band having same level of FL-378 was scored as B which indicated the homozygous allele of the donor parent for the particular SSR marker. However, heterozygous alleles were scored as H having both the bands of two parents. Score N indicated the absence of band. Importantly, heterozygous alleles always had the extra bands in most cases with bigger size. 3. Results and Discussion 3.1. Introgression of the saltol gene into Binadhan-7 using MAS The main aim of this study was to introgress the saltol gene into Binadhan-7 using a marker assisted backcrossing approach in order to
Alam et al. /The Agriculturists 10(2): 1-8 (2012) maximize the recovery of all of the desirable characteristics of the recurrent parent. During this process, further selection strategies associated with marker-assisted backcross breeding were done. The results of these selection strategies were as follows: 3.2. Confirmation of F1 plants DNA samples were collected from 10 F1 plants and PCR was carried out using one SSR marker RM21. PCR bands from all the 10 F1 plants were scored as H. Score H represented heterozygous alleles for donor and recipient parent. Fig. 1 shows the gel picture of F1 confirmation using a SSR marker RM21. All the F1s were confirmed as true F1 (Fig. 3). 3.3. Parental Survey A primer survey is an important prerequisite before starting marker-assisted backcross breeding. A comprehensive primer survey is required for successful foreground, recombinant and background selection. Polymorphic markers are essential for a MABC scheme. A marker which is monomorphic bears no value in selection work because this type of marker can not distinguish the two parental genotypes viz. Binadhan-7, the recurrent or recipient parent and FL-378, the donor parent of the MABC program. A total of 30 SSR primers were surveyed for finding out polymorphic markers and among them one primer was found as polymorphic. 3.4. Foreground selection at BC1F1 generation A total of 150 BC1F1 seeds were produced from 10 F1 plants (Table 3) and 105 plants were survived in the main fields. Foreground selection was performed on 72 plants. Foreground selection was carried out using a tightly linked salt tolerance marker RM21. The marker produced very conspicuous bands (Fig. 4, 5 & 6) and it was possible to identify the genetic constitution of the saltol QTLs very easily using agarose gel electrophoresis.
Marker-assisted selection for salt tolerant rice
Fig. 1. Marker-assisted foreground selection
Fig. 2. BC1F1 generation in the field
Fig. 3. Complete view of gel showing confirmation of F1s of Binadhan-7 x FL-378 using SSR marker RM 21
Table 1. Summary of microsatellite (SSR) marker used for BC1F1 DNA study Expected PCR product size (bp) 105 Annealing Temp. (oC) 55
Primer name
Chrom. position
Repeat Motif For.
Primer sequence ACAGTATTCCGTAGGCACGG GCTCCATGAGGGTGGTAGAG
RM 21
(TC)18
Rev.
Alam et al. /The Agriculturists 10(2): 1-8 (2012)
Table 2. Components of PCR cocktail (for 74 reactions) for foreground selection SL 1 2 3 4 5 6 Component 10X Buffer dNTPs Primer forward Primer reverse Taq polymerase ddH2O Total volume Quantity (for single reaction) 1.5 l 0.75 l 1.0 l 1.0 l 0.25 l 8.25 l Total 111.0l 55.5l 74.0l 74.0l 18.5l 610.5l 1554.0 l
Table 3. List of BC1F1 seeds produced SN 1 2 3 4 5 6 7 8 9 10 F1 1 2 3 4 5 6 7 8 9 10 Total Recurrent parent X Binadhan-7 X Binadhan-7 X Binadhan-7 X Binadhan-7 X Binadhan-7 X Binadhan-7 X Binadhan-7 X Binadhan-7 X Binadhan-7 X Binadhan-7 BC1F1 Seeds 20 14 19 9 17 15 8 11 16 21 150
Fig. 4. Gel picture of the foreground selection of BC1F1 population (1-28) with tightly linked salt tolerance marker RM21
Marker-assisted selection for salt tolerant rice
100 bp
Fig. 5. Gel picture of the foreground selection of BC1F1 population (29-58) with tightly linked salt tolerance marker RM21
100 bp
Fig. 6. Gel picture of the foreground selection of BC1F1 population (59-72) with tightly linked salt tolerance marker RM21 3.5. Foreground selection at BC1F1 generation with tightly linked marker RM21 Out of 72 plants, 33 plants were found showing the locus for tightly linked marker as heterozygous state (score H), 36 plants were found with the locus fixed for recipient allele (susceptible allele) (score A), 2 plants were found with the locus fixed for donor allele (resistant allele) (score B) and one score N indicated the absence of band. The two plants with B score were found may be due to accidental failure of backcrossing. The 33 plants with the H score for the tightly linked marker RM21 were subjected for recombinant selection. It was expected that those 33 individuals possessed the salt tolerant allele. So, these segregants were selected and promoted for further selection. The results were similar to the study of Neeraja et al. (2007) where foreground selection was performed with marker assisted backcrossing of rice for introgression of promising gene. Iftekharuddaula (2009) found similar results in his study on foreground selection in BC1F1 populations with marker assisted backcrossing to popular rice variety BR11.
8 4. Conclusions Based on the foreground selection with a tightly linked salt tolerance marker RM21, we selected 33 plants showing H scores were selected. The selected segregants were subjected to further recombinant and background selections in BC1F1 generation. And further foreground, recombinant and background selections will be done with appropriate markers up to BC3 or more for the development of salt tolerant rice lines and will be reported elsewhere. References ABSPII. 2006. Drought tolerant rice and salinity tolerance rice. Agricultural Biotechnology Support Project II- South Asia, 26 p. Bhuiyan, M. A. R. 2005. Efficiency in evaluating salt tolerance in rice using phenotypic and marker assisted selection. M.S. Thesis, Department of Genetics and Plant Breeding, Bangladesh Agricultural University, Mymensingh, Bangladesh, 96 p. Holland, J. B. 2004. Implementation of molecular markers for quantitative traits in breeding programs-challenges and opportunities. Proc. 4th Int. Crop Sci. Congress., Brisbane, Australia, 26 September-1 October, 2004. Hospital, F. and Charcosset A., 1997. Markerassisted introgression of quantitative trait loci. Genetics. 147:14691485. Iftekharuddaula, K. M. 2009. Comparison of new selection strategies of markerassisted backcrossing for a submergence tolerant gene in rice. Ph. D. dissertation.
Alam et al. /The Agriculturists 10(2): 1-8 (2012) Bangladesh agricultural Mymensingh. 75-91 p. University,
IPCC, 2007. Climate change in Asia too alarming to contemplate. Intergovernmental Panel on Climatic change, 85 p. Islam, M. M. 2004. Mapping salinity tolerance genes in rice (Oryza sativa L.) at reproductive stage. Ph. D. Dissertation. University of the Philippines Los Baos, College, Laguna, Philippines, 149 p. Neeraja, C. N., Maghirang-Rodriguez, R., Pamplona, A., Heuer, S., Collard, B. C. Y., Septiningsih, E. M., Vergara, G., Sanchez, D., Xu, K., Ismail, A. M., Mackill, D. J. 2007. A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theorical Application of Genetics, 115:767776. Niones, J. M. 2004. Fine mapping of the salinity tolerance gene on chromosome 1 of rice (Oryza sativa L.) using near-isogenic lines. M. S. dissertation. University of the Philippines Los Baos, College, Laguna, Philippines, 78 p. Thomson, M. J., Ismail, A. M., McCouch, S, R., Mackill, M. J. 2010. Marker Assisted Breeding (Chap. 20), Abiotic Stress Adaptation in Plants: Physiological, Molecular and Genomic Foundation, 451 469 pp. Zheng, K., Huang, N., Bennet, J. and Khush, G.S. 1995. PCR-based marker assisted selection in rice breeding. International Rice Research Institute, Los Baos, Laguna. Philippines, 24 p.
You might also like
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryFrom EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryRating: 3.5 out of 5 stars3.5/5 (231)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)From EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Rating: 4.5 out of 5 stars4.5/5 (121)
- Grit: The Power of Passion and PerseveranceFrom EverandGrit: The Power of Passion and PerseveranceRating: 4 out of 5 stars4/5 (588)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaFrom EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaRating: 4.5 out of 5 stars4.5/5 (266)
- The Little Book of Hygge: Danish Secrets to Happy LivingFrom EverandThe Little Book of Hygge: Danish Secrets to Happy LivingRating: 3.5 out of 5 stars3.5/5 (400)
- Never Split the Difference: Negotiating As If Your Life Depended On ItFrom EverandNever Split the Difference: Negotiating As If Your Life Depended On ItRating: 4.5 out of 5 stars4.5/5 (838)
- Shoe Dog: A Memoir by the Creator of NikeFrom EverandShoe Dog: A Memoir by the Creator of NikeRating: 4.5 out of 5 stars4.5/5 (537)
- The Emperor of All Maladies: A Biography of CancerFrom EverandThe Emperor of All Maladies: A Biography of CancerRating: 4.5 out of 5 stars4.5/5 (271)
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeFrom EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeRating: 4 out of 5 stars4/5 (5794)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyFrom EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyRating: 3.5 out of 5 stars3.5/5 (2259)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersFrom EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersRating: 4.5 out of 5 stars4.5/5 (344)
- Team of Rivals: The Political Genius of Abraham LincolnFrom EverandTeam of Rivals: The Political Genius of Abraham LincolnRating: 4.5 out of 5 stars4.5/5 (234)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreFrom EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreRating: 4 out of 5 stars4/5 (1090)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceFrom EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceRating: 4 out of 5 stars4/5 (895)
- Her Body and Other Parties: StoriesFrom EverandHer Body and Other Parties: StoriesRating: 4 out of 5 stars4/5 (821)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureFrom EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureRating: 4.5 out of 5 stars4.5/5 (474)
- The Unwinding: An Inner History of the New AmericaFrom EverandThe Unwinding: An Inner History of the New AmericaRating: 4 out of 5 stars4/5 (45)
- The Yellow House: A Memoir (2019 National Book Award Winner)From EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Rating: 4 out of 5 stars4/5 (98)
- On Fire: The (Burning) Case for a Green New DealFrom EverandOn Fire: The (Burning) Case for a Green New DealRating: 4 out of 5 stars4/5 (73)
- GRE Math FormulaDocument12 pagesGRE Math FormulaAnsuman Panda100% (2)
- Mendelian & Non-Mendelian GeneticsDocument37 pagesMendelian & Non-Mendelian GeneticsJubair Al-rashid100% (2)
- Introduction of Statistical GeneticsDocument61 pagesIntroduction of Statistical GeneticsJubair Al-rashid100% (2)
- Spectroscopic TechniqueDocument24 pagesSpectroscopic TechniqueJubair Al-rashidNo ratings yet
- Lactic Acid Bacteria Fundamentals and Practice PDFDocument536 pagesLactic Acid Bacteria Fundamentals and Practice PDFIrving Mayo100% (2)
- Marker-Assisted Backcrossing For Identification of Salt Tolerant Rice LinesDocument8 pagesMarker-Assisted Backcrossing For Identification of Salt Tolerant Rice LinesJubair Al-rashidNo ratings yet
- Mycorrhizal Symbioses How To Be Seen As A Good FungusDocument3 pagesMycorrhizal Symbioses How To Be Seen As A Good FungusJubair Al-rashidNo ratings yet
- Bioinformatics Lecture (Sequencing)Document33 pagesBioinformatics Lecture (Sequencing)Jubair Al-rashidNo ratings yet
- Genomic DNA Extraction From PlantDocument3 pagesGenomic DNA Extraction From Plantheroiszero2941No ratings yet
- Arbuscular Mycorrhiza Genetics of The Fungal PartnerDocument24 pagesArbuscular Mycorrhiza Genetics of The Fungal PartnerJubair Al-rashidNo ratings yet
- A Secreted Fungal Effector of Glomus Intraradices Promotes Symbiotic BiotrophyDocument6 pagesA Secreted Fungal Effector of Glomus Intraradices Promotes Symbiotic BiotrophyJubair Al-rashidNo ratings yet
- Plant Genomic DNA Extraction by CTAB - 2 - FionaDocument5 pagesPlant Genomic DNA Extraction by CTAB - 2 - FionayomnayasminNo ratings yet
- Chemical AnalysisDocument38 pagesChemical AnalysisJubair Al-rashidNo ratings yet
- MSTAT-C Manual Part1Document152 pagesMSTAT-C Manual Part1Jubair Al-rashid100% (1)
- Plabstat ManualDocument45 pagesPlabstat ManualJubair Al-rashidNo ratings yet
- MSTAT-C Manual Part3Document174 pagesMSTAT-C Manual Part3Jubair Al-rashid100% (1)
- MSTAT-C Manual Part2Document170 pagesMSTAT-C Manual Part2Jubair Al-rashid100% (1)
- MSTAT-C Manual Part1Document152 pagesMSTAT-C Manual Part1Jubair Al-rashid100% (1)
- IELTS Writing (Task 1 & 2)Document11 pagesIELTS Writing (Task 1 & 2)Jubair Al-rashid100% (1)
- Methods & Protocols For Arabidopsis Lipid AnalysesDocument28 pagesMethods & Protocols For Arabidopsis Lipid AnalysesJubair Al-rashidNo ratings yet
- Methods & Protocols For Arabidopsis Lipid AnalysesDocument28 pagesMethods & Protocols For Arabidopsis Lipid AnalysesJubair Al-rashidNo ratings yet
- Ham BrightDocument1 pageHam BrightJubair Al-rashidNo ratings yet
- Disaster Management Training PresentationDocument27 pagesDisaster Management Training PresentationJubair Al-rashidNo ratings yet
- Lamkey Ag Summit 2002Document35 pagesLamkey Ag Summit 2002Jubair Al-rashidNo ratings yet
- StrategyDocument1 pageStrategyJubair Al-rashidNo ratings yet
- Quantinova Sybr Green PCR Kit: ProfileDocument4 pagesQuantinova Sybr Green PCR Kit: ProfileRamakanth SatthenapalliNo ratings yet
- Isolation and Identification of Bacteria ThesisDocument7 pagesIsolation and Identification of Bacteria Thesisafbtbegxe100% (2)
- Marcelino, Christian - PCRDocument1 pageMarcelino, Christian - PCREhmMarcelinoNo ratings yet
- Surveillance For Probable Detection of Rabies Virus in Wild and Domestic AnimalsDocument4 pagesSurveillance For Probable Detection of Rabies Virus in Wild and Domestic AnimalszaheersbsNo ratings yet
- Food Chemistry: Andre Mu Ller, Hans SteinhartDocument9 pagesFood Chemistry: Andre Mu Ller, Hans SteinhartAlfredo CruzNo ratings yet
- Haidar Sitie Rafidah - 2006615010 - DNA FingerprintingDocument19 pagesHaidar Sitie Rafidah - 2006615010 - DNA FingerprintingHaidarNo ratings yet
- Bioknowledgy Quick Quiz On 3.5 Genetic Modification and BiotechnologyDocument5 pagesBioknowledgy Quick Quiz On 3.5 Genetic Modification and BiotechnologyPineappleHeadNo ratings yet
- Fastdigest Restriction Enzymes LabaidDocument2 pagesFastdigest Restriction Enzymes Labaidfnf102030No ratings yet
- Identification of Molecular Markers Associated With Genic Male Sterility in Tetraploid Cotton (Gossypium Hirsutum L.) Through Bulk Segregant Analysis Using A Cotton SNP 63K ArrayDocument7 pagesIdentification of Molecular Markers Associated With Genic Male Sterility in Tetraploid Cotton (Gossypium Hirsutum L.) Through Bulk Segregant Analysis Using A Cotton SNP 63K ArrayBlaxez YTNo ratings yet
- Sporothrix Brasiliensis and Feline Sporotrichosis in The Metropolitan Region of Rio de Janeiro, Brazil (1998-2018)Document14 pagesSporothrix Brasiliensis and Feline Sporotrichosis in The Metropolitan Region of Rio de Janeiro, Brazil (1998-2018)NealNo ratings yet
- MDRU Staff Recruitment - Final - 19 - 03 - 21 - NewDocument6 pagesMDRU Staff Recruitment - Final - 19 - 03 - 21 - Newjijesh jayadevanNo ratings yet
- CBC Diferenciação MatricalDocument12 pagesCBC Diferenciação MatricalliegedutraNo ratings yet
- BQ - MP - MahonDocument38 pagesBQ - MP - MahonAn K.No ratings yet
- (Pds Patklin) : Perhimpunan Dokter Spesialis Patologi Klinik Dan Kedokteran Laboratorium IndonesiaDocument13 pages(Pds Patklin) : Perhimpunan Dokter Spesialis Patologi Klinik Dan Kedokteran Laboratorium IndonesiaNida ChoerunnisaNo ratings yet
- CO010759qpcr HandbookDocument74 pagesCO010759qpcr HandbookCeciliaPistolNo ratings yet
- AIPMT - 2012 (Physics, Chemistry and Biology) Prelims Answer Key and Solution Code DDocument39 pagesAIPMT - 2012 (Physics, Chemistry and Biology) Prelims Answer Key and Solution Code DAnurag KasaudhanNo ratings yet
- Jurnal SweizerDocument17 pagesJurnal SweizerPanjiNo ratings yet
- A Comparative GC-MS Analysis of Bioactive Secondary Metabolites Produced by HalotolerantDocument16 pagesA Comparative GC-MS Analysis of Bioactive Secondary Metabolites Produced by HalotolerantAmineJaouedNo ratings yet
- January 2012 QP - Unit 4 Edexcel Biology A-LevelDocument24 pagesJanuary 2012 QP - Unit 4 Edexcel Biology A-LevelElissa MeflehNo ratings yet
- Clinical Manifestation, DiagnosisDocument13 pagesClinical Manifestation, DiagnosisGeorgiana BlagociNo ratings yet
- MolluscDocument25 pagesMolluscAisyah RamadaniNo ratings yet
- Molecular Biology Workflow Solutions 11 16Document40 pagesMolecular Biology Workflow Solutions 11 16Isaac Nicholas NotorioNo ratings yet
- Borzi 2018Document6 pagesBorzi 2018lokmenNo ratings yet
- GCE A - AS Level Biology A Topic Test - Genetics, Evolution and EcosystemsDocument29 pagesGCE A - AS Level Biology A Topic Test - Genetics, Evolution and Ecosystemsarfaat shahNo ratings yet
- BME 203 - Lecture No. 18-19Document45 pagesBME 203 - Lecture No. 18-19SagorNo ratings yet
- Jobm 201800252Document14 pagesJobm 201800252leilany casillasNo ratings yet
- Gene SilencingDocument7 pagesGene SilencingMaliha JahanNo ratings yet
- Primary Sample Collection Manual FinalDocument65 pagesPrimary Sample Collection Manual FinalUnihealth-Quezon LaboratoryNo ratings yet
- 69-RIAM FinalDocument3 pages69-RIAM FinalSoraya Eugenia Morales LópezNo ratings yet