Professional Documents
Culture Documents
CD4+ T უჯრედების მრავალფეროვნება
Uploaded by
EMD GROUPCopyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
CD4+ T უჯრედების მრავალფეროვნება
Uploaded by
EMD GROUPCopyright:
Available Formats
CD4 T-cell diversity
+
John O'Shea, Arian Laurence and Adewole Adamson
CD4+ T helper (TH) cells are central players in orchestrating innate and adaptive immune responses. TH cells have been classically considered to belong to one of two subsets TH1 cells or TH2 cells each of which has unique cytokine products, signalling pathways and lineage-specific transcription factors or master regulators. Recently, TH cells that secrete IL-17 (TH17 cells) have emerged as a third lineage of CD4+ TH cells. Together with regulatory T (TReg) cells, which preserve peripheral tolerance, these two newly described Tcell subsets have raised fundamental questions about lineage commitment and fate determination of CD4+ T cells. This Poster depicts the various cytokines, transcription factors and signalling pathways that are associated with the differentiation, survival and function of CD4+ effector T cells. The differentiation of CD4+ Tcells is typically depicted as a one-way route, implying that there is terminal differentiation with little plasticity in cytokine responses. However, recent data argue for more complexity and flexibility than was previously assumed. As with any model, this is a work in progress and subject to enhancement in the coming years.
IMMUNOLOGY
IL-4 IFN, TGF STAT6 GATA3 GATA3 IFN STAT4 IL-12 IL-21 IL-1 IL-2 STAT5 SMAD FOXP3 IL-23 IL-18 RUNX1 STAT3 TNF SMAD IRF4 AHR RUNX1 TGF SMAD FOXP3 IL-17 SOCS3, STAT1, STAT5, FOXP3 IL-23 RORt ROR STAT3 IL-17 RUNX3 TGF, IL-4, IL-21 IFN, IL-2, IL-4, IL-12, IL-27, RA Notch HLX T-bet STAT1 IL-4 STAT1 T-bet GATA3 IL-12 IFN
Naive CD4+ T cell
Antigen-presenting cell TGF RA IL-6, IL-21 IFN IL-6
IL-4 IL-25
STAT6 Notch
IRF4 MAF
JUNB
IL-4, IL-5, IL-10, IL-13, IL-21, IL-31 Humoral immunity, allergy and other functions
TH2 cell
T-bet STAT4
IFN, TNF, lymphotoxin, IL-10 Cell-mediated immunity and type I hypersensitivity
IFNproducing TH17 cell
TH1 cell
Subset flexibility and plasticity
TH17 cell
IL-6 TGF
RORt IL-4, IFN IL-23 IL-6
Inducible TReg cell
IL-17A, IL-17F, IL-21, IL-22 Inflammation and pathogen defence
TGF, IL-10, IL-35 Self tolerance and immune suppression
IL-10producing TH17 cell
IL-10, IL-17A, IL-17F
IL-22producing TH17 cell
IL-21, IL-22, IL-17A, IL-17F, IL-26, CCL20
At present, lineage commitment is defined by the signature cytokines that differentiated cells secrete IFN, IL-17 (IFN, IL-4 and IL-17 for TH1, TH2 and TH17 cells, respectively). However, additional complexity is becoming evident. For example, although TReg cells are thought to be immunosuppressive, all CD4+ T-cell subsets can produce the immunoregulatory cytokine IL-10, which is crucial for dampening immune responses. Furthermore, TH17 cells can also produce IFN, and so the relationship between TH1 cells and TH17 cells is currently being intensively scrutinized. Similarly, although TH17 cells produce IL-21 and IL-22, it also seems that under some circumstances, T cells can produce IL-21 or IL-22 without producing IL-17 (not shown). Another complication is that models that imply the selective expression of master regulators might not be appropriate. For example, FOXP3 and RORt can be co-expressed and can interact. Functionally, it seems that some TReg cells can be induced to become TH17 cells. So, an important issue in the field is whether each CD4+ T-cell subset should be viewed as a terminally differentiated lineage or whether the subsets retain some flexibility. The extent to which some aspects of T-cell subsets are firmly fixed and others remain plastic is the subject of intense investigation.
TH1 cell
IL-12 IL-12R1 IL-12R2 IFNGR1
IFN IFNGR2
Notch ligand
IL-4 Notch receptor IL-4R c
IL-2 c
TH2 cell
IL-2RB IL-2RA
CD4+ T-cell differentiation
Following contact with antigen-presenting cells (APCs), signals generated by the T-cell receptor (TCR) and co-stimulatory molecules initiate the process by which naive CD4+ T cells begin to differentiate towards one of several fates. In the context of an appropriate signal through the TCR, the cytokine milieu that is generated by APCs is an important factor that influences differentiation. IL-12 activates STAT4 and drives naive CD4+ T cells to become TH1 cells, which produce IFN. Signals from the TCR, as well as from IL-12 and IFN (acting through STAT4 and STAT1, respectively), increase the expression of the transcription factor T-bet, which promotes IFN production and commitment to the TH1-cell lineage. TH1 cells are important for host defence against intracellular bacteria. Naive CD4+ T cells are induced to become TH2 cells through the secretion of IL-4 by innate immune cells, which signals through STAT6. This leads to expression of the transcription factor GATA3, in turn resulting in the production of IL-4, IL-5 and IL-13, which are important for host defence against helminths and contribute to the pathogenesis of asthma and allergy. TReg cells can develop from thymic CD4+ T-cell precursors in the presence of TGF and IL-2. These are termed natural TReg cells (not shown). In the periphery, naive CD4+ T cells can also be converted to become inducible TReg cells by signalling through STAT5 in the presence of TGF, which results in upregulation of the transcription factor FOXP3. TReg cells secrete low levels of IL-2 and IFN, and instead they produce high levels of IL-10, IL-35 and TGF. Retinoic acids, which are abundant in the liver and intestine, increase FOXP3 expression. TReg cells have an important role in peripheral self tolerance and immune suppression. TH17 cells develop from naive CD4+ T cells in response to IL-6, IL-21, TGF and IL-1b. IL-6 and IL-21 activate STAT3, which increases the expression of the transcription factors RORt and ROR, which in turn promote the expression of IL-17A, IL-17F, IL-21 and IL-22. IL-23 seems to stabilize and increase the pathogenicity of TH17 cells. TH17 cells are important for host defence against extracellular bacteria and are involved in mediating autoimmune disease.
Cytoplasm STAT6 STAT4
P P P P P
STAT5
STAT1
Intracellular Notch Notch3 Notch1, Notch2
STAT1
STAT6
Nucleus STAT4
P
MAML RBPJ
P
STAT5
GATA3
P
HLX
NFB
MAML RBPJ T-bet T-bet
GATA3 GATA3, GFI1
MAML RBPJ MAF NFAT2 AP1
IFN, TNF, lymphotoxin
IL-4, IL-5, IL-13
Inducible TReg cell
AHR ligand Retinoic acid c
IL-2
TGF IL-2RB TGFR
IL-6 IL-6ST R-SMAD IL-6R
IL-21 IL-27 c IL-21R IL-6ST IL-27RA IL-1RAP
TH17 cell
IL-1 IL-1R1
IL-2RA
STAT5
STAT3
Cytoplasm AHR HSP90
R-SMAD Co-SMAD ?
SOCS3 STAT1 RAR
P P
STAT5
FOXP3
R-SMAD Co-SMAD FOXP3
STAT3
RUNX1
STAT3?
Nucleus
RAR
AHR ? ARNT ?
P
RUNX1
RUNX1 RORt ROR
TGF, IL-10, IL-35
ETS1
RORt, ROR
IL-17A, IL-17F, IL-21, IL-22
c, common cytokine receptor -chain; AHR, aryl hydrocarbon receptor; AP1, activator protein 1; ARNT, aryl receptor nuclear transporter; CCL20, CC-chemokine ligand 20; Co-SMAD, co-mediator SMAD; FOXP3, forkhead box P3; GATA3, GATA-binding protein 3; GFI1, growth-factor independent 1; HLX, H2.0-like homeobox 1; HSP90, heat-shock protein 90; IFN, interferon-; IL, interleukin; IL-1RAP, IL-1 receptor accessory protein; IL-6ST, IL-6 signal transducer; IRF, interferon-regulatory factor; MAML, Mastermind-like; NFAT, nuclear factor of activated Tcells; NF-B, nuclear factor-B; RA, retinoic acid; RAR, retinoic acid receptor; RBPJ, recombination-signal-binding protein for immunoglobulin- J region (also known as CSL); ROR, retinoic-acid-receptor-related orphan receptor; R-SMAD, receptor-regulated SMAD; RUNX, Runt-related transcription factor; SMAD, mothers against decapentaplegic homologue; SOCS3, suppressor of cytokine signalling 3; STAT, signal transducer and activator of transcription; TGF, transforming growth factor-; TNF, tumour-necrosis factor.
Abbreviations
Affiliations
John OShea, Arian Laurence and Adewole Adamson are at the Molecular Immunology and Inflammation Branch, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, Maryland 20892, USA. e-mail: osheaj@arb.niams.nih.gov Edited by Lucy Bird and Kirsty Minton; copy edited by Rachel David; designed by Simon Bradbrook.
UCB www.ucb-group.com
With a unique combination of expertise in biology and chemistry, UCB is a global biopharmaceutical company focusing on severe diseases in two main therapeutic areas CNS and Autoimmune and Inflammatory diseases. UCB operates in more than 40 countries with a global team of Research, Development and Commercial operations. UCB is focused on the needs of patients as well as bringing effective treatments to them using both novel biological entities (NBEs) and novel chemical entities (NCEs). These treatments include Cimzia, a Pegylated Fab antibody fragment targeted at Crohns disease and rheumatoid arthritis, and NCEs, such as Xyzal, Keppraand Neupro. In addition to being a world leader in allergy and epilepsy, withmore than 10 registered drugs for the treatment of these conditions, UCB also has a broad pipeline of both NBE and NCE therapies in clinical development with the potential to treat many chronic inflammatory diseases. By integrating biology and chemistry, UCB can gain much deeper insights into disease pathways as well as producing the optimum therapeutic for each target. UCB's pioneering A2HiT project, which is utilizing leadingantibody research to design more effective NCEs, is one of the most exciting examples of how UCB is capitalizing on thesynergies betweenbiology and chemistry as the company moves forward to develop new drugs for the 21st century.
You might also like
- Textbook of Clinical NeuropsychiatryDocument746 pagesTextbook of Clinical Neuropsychiatrymayamanush100% (1)
- Advanced Guide For Professional Homeopaths Luc de Schepper.04253 1Document24 pagesAdvanced Guide For Professional Homeopaths Luc de Schepper.04253 1bshowalter_1No ratings yet
- Zhang 2018Document37 pagesZhang 2018twarrenNo ratings yet
- To Study The Role of T-Regulatory Cells in The Infectious and Non-Infectious UveitisDocument30 pagesTo Study The Role of T-Regulatory Cells in The Infectious and Non-Infectious UveitisRavi SharmaNo ratings yet
- Il 17Document25 pagesIl 17mireya nuñezNo ratings yet
- Poblaciones LT PlasticidadDocument6 pagesPoblaciones LT PlasticidadLeidy Nayerli Garcia RodriguezNo ratings yet
- Cytokines and Thelper Subsets: MCB 150, PR CoscoyDocument42 pagesCytokines and Thelper Subsets: MCB 150, PR CoscoyMac Kevin MandapNo ratings yet
- Clinical Outlook For Type 1 and Foxp3 T Regulatory Cell Based TherapyDocument8 pagesClinical Outlook For Type 1 and Foxp3 T Regulatory Cell Based TherapyAgustin MendezNo ratings yet
- Cheng Et Al 2006 - Act - Il-17rDocument5 pagesCheng Et Al 2006 - Act - Il-17rNilabh RanjanNo ratings yet
- LPS-TLR4 Tranduction PathwaysDocument7 pagesLPS-TLR4 Tranduction PathwaysGERMAN MATIAS TAPIA CURIMILNo ratings yet
- PosterDocument1 pagePostermomenajib100% (1)
- Benevides2018 PDFDocument15 pagesBenevides2018 PDFNayche TortatoNo ratings yet
- 2001 - Regulation of IFN-γ signaling is essential for the cytotoxic activity of CD8+Document10 pages2001 - Regulation of IFN-γ signaling is essential for the cytotoxic activity of CD8+顏世隆No ratings yet
- Interleukin-17 and Its Expanding Biological Functions: ReviewDocument11 pagesInterleukin-17 and Its Expanding Biological Functions: ReviewanhiramdhaniNo ratings yet
- Dejaco-Et-Al 06 - (Imbalance of Regulatory T Cells in Human Autoimmune Diseases)Document12 pagesDejaco-Et-Al 06 - (Imbalance of Regulatory T Cells in Human Autoimmune Diseases)Luana DiEmmeNo ratings yet
- Cytolytic CD4+ T Cells inDocument11 pagesCytolytic CD4+ T Cells inFrancisco Ibañez IrribarraNo ratings yet
- STAT3 Regulates Cytokine-Mediated Generation of Inflammatory Helper T CellsDocument6 pagesSTAT3 Regulates Cytokine-Mediated Generation of Inflammatory Helper T Cellssa571922No ratings yet
- Biology: Programmed Cell Death 1 Ligand (PD-L1) On T Cells Generates Treg Suppression From MemoryDocument4 pagesBiology: Programmed Cell Death 1 Ligand (PD-L1) On T Cells Generates Treg Suppression From MemoryMarlyNationTVNo ratings yet
- Th17 Cells: The Complete Solution For Th17 Cell AnalysisDocument4 pagesTh17 Cells: The Complete Solution For Th17 Cell AnalysisEMD GROUPNo ratings yet
- J Coi 2007 04 005Document6 pagesJ Coi 2007 04 005Jayanth BalakuntlaNo ratings yet
- Cytokine Biology - Fish Lectures: Cytokines and Their Role in The Immune SystemDocument12 pagesCytokine Biology - Fish Lectures: Cytokines and Their Role in The Immune SystemChristine QianNo ratings yet
- Th17 Cells InflamationDocument4 pagesTh17 Cells InflamationJayanth BalakuntlaNo ratings yet
- Citocinas ReviewDocument91 pagesCitocinas ReviewHugo SantanaNo ratings yet
- Perspectives: Diversification of T-Helper-Cell Lineages: Finding The Family Root of IL-17-producing CellsDocument5 pagesPerspectives: Diversification of T-Helper-Cell Lineages: Finding The Family Root of IL-17-producing CellsIván DiazNo ratings yet
- Table 10-1. Major Properties of Human Interleukins. Interleukin Principal Cell Source Principal EffectsDocument8 pagesTable 10-1. Major Properties of Human Interleukins. Interleukin Principal Cell Source Principal EffectsKhrisnanto NugrohoNo ratings yet
- 2008 Good Good Clonal Regulatory T Cells Specific For A Red Blood Cell Auto Antigen in Human Autoimmune Hemolytic AnemiaDocument9 pages2008 Good Good Clonal Regulatory T Cells Specific For A Red Blood Cell Auto Antigen in Human Autoimmune Hemolytic AnemiaNguyen Tien HuyNo ratings yet
- Molecular Mechanisms of T Helper Cell Differentiation and Functional SpecializationDocument15 pagesMolecular Mechanisms of T Helper Cell Differentiation and Functional Specializationhendarto ArifNo ratings yet
- Mechanisms of Allergic Immunity: Crah1@le - Ac.ukDocument98 pagesMechanisms of Allergic Immunity: Crah1@le - Ac.ukNaba ManalNo ratings yet
- TregsDocument1 pageTregsAndreaNo ratings yet
- Prolactin Increase Frecuency of T Helper FolicularDocument15 pagesProlactin Increase Frecuency of T Helper Folicularauroragamez2015No ratings yet
- Hydrolysis of InterleukinDocument12 pagesHydrolysis of InterleukinRindi GurciNo ratings yet
- The TLR Signaling Adaptor TRAM Interacts With TRAF6 To Mediate Activation of The Inflammatory Response by TLR4Document21 pagesThe TLR Signaling Adaptor TRAM Interacts With TRAF6 To Mediate Activation of The Inflammatory Response by TLR4Nadia NasrNo ratings yet
- Jurnal DafiqDocument5 pagesJurnal DafiqDafiq Mihal F YusufNo ratings yet
- Collins 2008 IL-17 in Inflammation and AutoimmunityDocument16 pagesCollins 2008 IL-17 in Inflammation and AutoimmunityNilabh RanjanNo ratings yet
- Activation of T Lymphocytes: Hypersensitivity Referring To Tissue Damage Caused by An ImmuneDocument8 pagesActivation of T Lymphocytes: Hypersensitivity Referring To Tissue Damage Caused by An ImmuneJorge SalinasNo ratings yet
- Cooke Short Review 081606 Web 2Document4 pagesCooke Short Review 081606 Web 2Thays DornellesNo ratings yet
- Interleukin-33 in Asthma How Big of A Role Does It PlayDocument8 pagesInterleukin-33 in Asthma How Big of A Role Does It PlayFabiula AbreuNo ratings yet
- Regulatory T Cell I AtheroschlerisisDocument13 pagesRegulatory T Cell I AtheroschlerisisganangahimsaNo ratings yet
- Activation of CD8 T Cells Induces Expression of CD4, Which Functions As A Chemotactic ReceptorDocument6 pagesActivation of CD8 T Cells Induces Expression of CD4, Which Functions As A Chemotactic ReceptorKevin MaiseyNo ratings yet
- Wiki T CellDocument12 pagesWiki T Cellالولد الخطير تابعونيNo ratings yet
- 2022 Article 210Document13 pages2022 Article 210uttamNo ratings yet
- Lymphokines & Cytokines: Immunology & DiseaseDocument34 pagesLymphokines & Cytokines: Immunology & DiseasejaneNo ratings yet
- AsthmaDocument100 pagesAsthmaMohamed HefnyNo ratings yet
- Shimizu 1997Document11 pagesShimizu 1997Araceli Enríquez OvandoNo ratings yet
- Article Review MICR7002 FinalDocument8 pagesArticle Review MICR7002 FinalMiranda KeikuNo ratings yet
- Cell-Mediated ImmunityDocument15 pagesCell-Mediated ImmunityFatema Al-KananiNo ratings yet
- Serody 2012Document6 pagesSerody 2012bixagif369No ratings yet
- Gervaziev 2010Document8 pagesGervaziev 2010Ludmila OleninaNo ratings yet
- Slide Jurnal 1Document19 pagesSlide Jurnal 1Benni Andica SuryaNo ratings yet
- R 2006 - Regulatory T Cells, Tumour ImmunityDocument13 pagesR 2006 - Regulatory T Cells, Tumour Immunity1262615286No ratings yet
- Nihms389960 PDFDocument12 pagesNihms389960 PDFLuis TolentinoNo ratings yet
- Colesterol: Función Biológica e Implicaciones MédicasDocument5 pagesColesterol: Función Biológica e Implicaciones MédicasJuan manuel jiménez estradaNo ratings yet
- Regulatory T-Cell Suppressor Program Co-Opts TransDocument16 pagesRegulatory T-Cell Suppressor Program Co-Opts Transygilad9139No ratings yet
- TNF Ligands and Receptors A Matter of Life and Death: ReviewDocument21 pagesTNF Ligands and Receptors A Matter of Life and Death: ReviewLizeth Rincon DelgadoNo ratings yet
- Mosser, Justin XiaDocument10 pagesMosser, Justin XiaIzabela HondiakoviskyNo ratings yet
- The Role of IL-17 and Related Cytokines in Inflammatory Autoimmune DiseaseDocument12 pagesThe Role of IL-17 and Related Cytokines in Inflammatory Autoimmune DiseaserodtobingNo ratings yet
- C04 Innate ImmunityDocument31 pagesC04 Innate ImmunityforeveraldyNo ratings yet
- T Cells Development: Selda S. Ezzel-DeenDocument67 pagesT Cells Development: Selda S. Ezzel-DeenTamara ElyasNo ratings yet
- Protective Roles of Mast Cells Against Enterobacterial Infection Are Mediated by Toll-Like Receptor 4Document7 pagesProtective Roles of Mast Cells Against Enterobacterial Infection Are Mediated by Toll-Like Receptor 4harumiaikoNo ratings yet
- 10 1016@j TCB 2020 06 003Document10 pages10 1016@j TCB 2020 06 003Maria Aponte RuizNo ratings yet
- Co-signal Molecules in T Cell Activation: Immune Regulation in Health and DiseaseFrom EverandCo-signal Molecules in T Cell Activation: Immune Regulation in Health and DiseaseMiyuki AzumaNo ratings yet
- IITC Life Science - Certificate of RepresentationDocument1 pageIITC Life Science - Certificate of RepresentationEMD GROUPNo ratings yet
- პროტეასომის ინჰიბიცია კიბოს მკურნალობაშიDocument1 pageპროტეასომის ინჰიბიცია კიბოს მკურნალობაშიEMD GROUPNo ratings yet
- ტერატომებიდან ემბრიონულ ღეროვან უჯრედებამდე - პლურიპოტენტურობის აღმოჩენაDocument1 pageტერატომებიდან ემბრიონულ ღეროვან უჯრედებამდე - პლურიპოტენტურობის აღმოჩენაEMD GROUPNo ratings yet
- Bibby Scientific LTD - Authorisation LetterDocument1 pageBibby Scientific LTD - Authorisation LetterEMD GROUPNo ratings yet
- C ჰეპატიტის ვირუსის რეპლიკაციის დათრგუნვის გზებიDocument2 pagesC ჰეპატიტის ვირუსის რეპლიკაციის დათრგუნვის გზებიEMD GROUPNo ratings yet
- მეზენქიმური ღეროვანი უჯრედებიDocument1 pageმეზენქიმური ღეროვანი უჯრედებიEMD GROUPNo ratings yet
- საკვების მიღებისა და ენერგეტიკული ბალანსის ნერვული რეგულაციაDocument1 pageსაკვების მიღებისა და ენერგეტიკული ბალანსის ნერვული რეგულაციაEMD GROUPNo ratings yet
- იმუნური სისტემა კიბოს წინააღმდეგDocument1 pageიმუნური სისტემა კიბოს წინააღმდეგEMD GROUPNo ratings yet
- II ტიპის შაქრიანი დიაბეტიDocument1 pageII ტიპის შაქრიანი დიაბეტიEMD GROUP100% (1)
- ბეტა ამილოიდი და ტაუ ცილა ალცჰაიმერის დაავადების პათოგენეზშიDocument1 pageბეტა ამილოიდი და ტაუ ცილა ალცჰაიმერის დაავადების პათოგენეზშიEMD GROUPNo ratings yet
- დნმ-ის დაზიანების მექანიზმები კანცეროგენეზისა და კიბოს მკურნალობის დროსDocument1 pageდნმ-ის დაზიანების მექანიზმები კანცეროგენეზისა და კიბოს მკურნალობის დროსEMD GROUPNo ratings yet
- I ტიპის შაქრიანი დიაბეტიDocument1 pageI ტიპის შაქრიანი დიაბეტიEMD GROUPNo ratings yet
- ანტიგენების პროცესინგი და პრეზენტაციაDocument1 pageანტიგენების პროცესინგი და პრეზენტაციაEMD GROUPNo ratings yet
- მიტოქონდრიის ფუნქცია და კიბოDocument1 pageმიტოქონდრიის ფუნქცია და კიბოEMD GROUPNo ratings yet
- Toll-ის მსგავსი რეცეპტორებიDocument4 pagesToll-ის მსგავსი რეცეპტორებიEMD GROUPNo ratings yet
- აივ ვირუსის საწინააღმდეგო იმუნური პასუხიDocument1 pageაივ ვირუსის საწინააღმდეგო იმუნური პასუხიEMD GROUPNo ratings yet
- უჯრედების იმუნომაგნიტური სეპარაციაDocument8 pagesუჯრედების იმუნომაგნიტური სეპარაციაEMD GROUPNo ratings yet
- აპოფტოზის მოლეკულური მექანიზმებიDocument1 pageაპოფტოზის მოლეკულური მექანიზმებიEMD GROUPNo ratings yet
- NK და NKT უჯრედების მარკერებიDocument2 pagesNK და NKT უჯრედების მარკერებიEMD GROUPNo ratings yet
- ადამიანის CD ანტიგენებიDocument1 pageადამიანის CD ანტიგენებიEMD GROUP100% (1)
- Th17 Cells: The Complete Solution For Th17 Cell AnalysisDocument4 pagesTh17 Cells: The Complete Solution For Th17 Cell AnalysisEMD GROUPNo ratings yet
- 4 Lab ManualDocument160 pages4 Lab ManualAziz EzoNo ratings yet
- Portfolio Clinical Case Study 2Document29 pagesPortfolio Clinical Case Study 2api-277136509No ratings yet
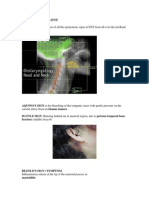
- Eponymous Signs in ENTDocument11 pagesEponymous Signs in ENTDrTarek Mahmoud Abo KammerNo ratings yet
- Gastric VolvulusDocument6 pagesGastric VolvulusIosif SzantoNo ratings yet
- Chap 11 ForthcomingDocument61 pagesChap 11 ForthcomingAna RebeloNo ratings yet
- Efusi Pleura Ec Ca ParuDocument33 pagesEfusi Pleura Ec Ca ParuRahma Putri KinasihNo ratings yet
- ADC W2016 Damir TanyaDocument50 pagesADC W2016 Damir TanyaThamadapu Swathi100% (1)
- ESR Ebook For Undergraduate Education in Radiology - 04 Breast ImagingDocument186 pagesESR Ebook For Undergraduate Education in Radiology - 04 Breast ImagingmarysrbNo ratings yet
- Texas Children's Hospital 5 Annual Advanced Practice Provider ConferenceDocument64 pagesTexas Children's Hospital 5 Annual Advanced Practice Provider ConferenceEngSabbaghNo ratings yet
- Total Parenteral Nutrition: The Long and The Short of It: Duna PennDocument7 pagesTotal Parenteral Nutrition: The Long and The Short of It: Duna PennDsk IndryNo ratings yet
- Lesson: Dr. Leon Hammer, M.DDocument14 pagesLesson: Dr. Leon Hammer, M.DRocío JMNo ratings yet
- Nature Magazine - 14 January 2016Document290 pagesNature Magazine - 14 January 2016David Košić100% (1)
- 1 s2.0 S1040842801002141 MainDocument22 pages1 s2.0 S1040842801002141 MainHayat BouchoumNo ratings yet
- 3M Material Safety Data Sheet Electrical Moisture Sealant Pads (Ems Ii)Document7 pages3M Material Safety Data Sheet Electrical Moisture Sealant Pads (Ems Ii)danielliram993No ratings yet
- Examining The PrecordiumDocument83 pagesExamining The PrecordiumnicolNo ratings yet
- Radiography in Osteoporosis: Sivasubramanian Srinivasan and Wilfred C. G. PehDocument16 pagesRadiography in Osteoporosis: Sivasubramanian Srinivasan and Wilfred C. G. PehAdriana NavaNo ratings yet
- 2022 2oENEM 1oDIA CMTDDocument95 pages2022 2oENEM 1oDIA CMTDjulianaaragamenoNo ratings yet
- Vihita Chemicals CaseDocument12 pagesVihita Chemicals Casesanju100% (2)
- A Date With DestinyDocument27 pagesA Date With DestinyJayvee Fulgencio100% (2)
- E WasteDocument64 pagesE WasteKartik MuppirisettyNo ratings yet
- CancerDocument23 pagesCancerpitkoliNo ratings yet
- Alopecia Unapproved Treatmentes or IndicationsDocument10 pagesAlopecia Unapproved Treatmentes or IndicationsdanizituNo ratings yet
- Month by MonthDocument28 pagesMonth by MonthChoi Han KimNo ratings yet
- Intl Journal of Cancer - 2024 - Oyelere - Coffee Consumption Is Associated With A Reduced Risk of Colorectal CancerDocument10 pagesIntl Journal of Cancer - 2024 - Oyelere - Coffee Consumption Is Associated With A Reduced Risk of Colorectal CancerRoel PlmrsNo ratings yet
- Free Radical TestDocument2 pagesFree Radical TestMr. Pushparaj GhodkeNo ratings yet
- OSHAD-SF - TG - New and Expectant Mothers - A Guide For Employers - V3.1 EnglishDocument30 pagesOSHAD-SF - TG - New and Expectant Mothers - A Guide For Employers - V3.1 EnglishNiel Brian VillarazoNo ratings yet
- Grandi 2019. Hormonal Contraception in Women With Endometriosis - A Systematic ReviewDocument11 pagesGrandi 2019. Hormonal Contraception in Women With Endometriosis - A Systematic ReviewcespersiNo ratings yet
- Spondilitis TBDocument28 pagesSpondilitis TBIdza Fariha AfriNo ratings yet