Professional Documents
Culture Documents
Myxoid Liposarcoma
Uploaded by
Sonny Dizon PareñasOriginal Description:
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Myxoid Liposarcoma
Uploaded by
Sonny Dizon PareñasCopyright:
Available Formats
Soft Tissue Tumors: Liposarcoma: Myxoid liposarcoma
Identity
Soft Tissue Tumors:Liposarcoma:Myxoid liposarcoma Phylum Other names Myxoid-round cell liposarcoma Sarcomas are relatively rare malignant tumours and comprise less than 10% of all Note cancers. Classical classifications of sarcomas are based on the site of tumour (bone or soft tissue). Soft tissue sarcoma (STS) is the collective term used for malignancies arising in muscles, fat, vessels, the peripheral nervous system and fibrous tissue. Histopathologic examination of such tumours has revealed a large number of distinct entities, each displaying its own morphologic and clinical characteristics. Cytogenetic and molecular genetic analyses have shown that some of these STS are characterized by specific chromosomal translocations, whereas other STS show complex genetic aberrations. Liposarcoma is the most common soft tissue malignancy in adults accounting for at least 20% of all sarcomas in this age group. Myxoid-round cell liposarcoma is a subtype of liposarcoma characterized by the presence of the reciprocal chromosomal translocation t(12;16)(q13;p11). This translocation creates the FUSDDIT3 chimeric gene.
Classification
Liposarcoma is a lipogenic tumour subclassified into four main histologic groups, including well-differentiated liposarcoma (lipoma-like and sclerosing types), myxoid-round cell liposarcoma, pleomorphic liposarcoma, and dedifferentiated liposarcoma. The histologic group is predictive of both the clinical course of the disease and the ultimate prognosis.
Clinics and Pathology
Cytogenetics
Cytogenetics Cytogenetics analyses have shown that several lipogenic tumours are Morphological characterized by specific chromosomal abnormalities, the best known was the reciprocal translocation t(12;16)(q13;p11) of myxoid-round cell liposarcoma,
described about twenty years ago.This translocation results in a fusion gene consisting of the 5' part of the FUS (TLS) gene and the complete coding region of the CHOP gene (see fig.1).
Genes involved and Proteins
Gene Name Location Dna / Rna FUS (TLS) 16p11 The FUS gene consists of 15 exons located within 11 kb of genomic DNA, and the exon 1 contains a 72-bp untranslated region and the translation initiation codon. The location of the FUS gene was identified as 16p11 by the site of the breakpoint in the translocation. The assignment was further narrowed to 16p11.2 by cytogenetic studies. FUS is rearranged in myxoid liposarcomas in the characteristic chromosomal translocation t(12;16)(q13;p11). The FUS protein, provisionally designated TLS (translocated in liposarcoma), and then called FUS, contains an RNA-recognition motif and is a component of nuclear riboprotein complexes. Lack of FUS in mice causes lethallity into neonatal period, it influences lymphocyte development in a non-cell-intrinsic manner, it has an intrinsic role in the proliferative responses of B cells to specific mitogenic stimuli, and it is required for the maintenance of genomic stability. The involvement of a nuclear riboprotein in these processes in vivo indicates that FUS is important in genome maintenance. Variants: FUS has been also shown a partner of gene fusions linked in other malignances: fused to ERG in acute myeloid leukaemia with t(16:21)(p11;q22), fused to CREB3L2 in low-grade fibromyxoid .sarcoma (LGFMS) by a translocation between chromosome bands 7q33-q34 (CREB3L2) and 16p11 (FUS) or fused to ATF1 in histiocytoma.
Protein
Somatic mutation
Gene Name Location Dna / Rna
Protein
DDIT3 (CHOP) 12q13 The DDIT3 gene was isolated from human cells and has a high level of conservation with previously described hamster gene. Each is composed of 4 exons with intron/exon junctions maintained at identical positions. They showed 91% identity in amino acid sequence and 78% identity in nucleotide sequence. The gene is located on chromosome 12 (12q13.1-q13.2) CHOP (C/EBP-homologous protein) is a nuclear protein which was identified as a dominant-negative inhibitor of the transcription factors C/EBP and LAP. The protein also was called DDIT3 for DNA damage-inducible transcript 3' and GADD153 for 'growth arrest- and DNA damage-inducible gene. DDIT3 is consistently rearranged in myxoid liposarcomas in the characteristic chromosomal translocation t(12;16)(q13;p11). Its molecular characterization showed that the DDIT3 gene is fused with a gene on chromosome 16 named FUS.
Somatic mutation
Variants: An analysis of peripheral blood samples from 19 patients with myxoid liposarcoma linked to t(12;16) and from 1 patient with myxoid liposarcoma associated to t(12;22;20) chromosomal translocation, resulting in the fusion of the DDIT3 and EWS genes, found FUS-DDIT3 hybrid fragments in 3 patients with t(12;16) and the EWS-DDIT3 hybrid in the patient with the latter translocation.
Result of the chromosomal anomaly
Hybrid Gene
The FUS-CHOP fusion genes consist of the 5 promoter region and exons 1-5 or, more rarely, 1-7 or 1-8 of FUS gene fused to the complete coding region, including exons 1-4 or 2-4, of CHOP (DDIT3) gene.
Fusion Protein Description
Myxoid liposarcoma tumor developed in a FUS-DDIT3 transgenic mice (40X objetive, H-E staining).
Oncogenesis Oncogenic properties Transcriptional control of the fusion gene is dominated by the FUS housekeeping type of regulatory region, leading to stable expression of the fusion protein in tumor cells. The transforming properties of the FUS-DDIT3 fusion protein have been demonstrated in NIH 3T3 cells and fibroblasts. In the FUS-DDIT3 fusion, transcriptional activation is specifically conferred on the chimeric protein by the FUS segment after the translocation event. The portion of FUS that is present in the FUS-DDIT3 and FUS-ERG fusion proteins is similar
and this part has been shown to be an autonomous transcriptional activation domain. The protein most likely functions as an abnormal transcription factor acting on a number of downstream target genes. Mouse models In vivo, mice expressing FUS-DDIT3 develop liposarcomas. Overexpression of FUS-DDIT3 transgene driven by the elongation factor 1alpha (EF1alpha) promoter to all tissues, results in most of the symptoms of human liposarcomas, including the presence of lipoblasts with round nuclei, accumulation of intracellular lipid, induction of adipocyte-specific genes and a concordant block in the differentiation program (see figure 2). No tumours of other tissues were found in these transgenic mice despite widespread activity of the EF1alpha promoter. This establishes FUS-DDIT3 overexpression as a key determinant of human liposarcomas and provided the first in vivo evidence for a link between a fusion gene created by a chromosomal translocation and a solid tumour. In contrast, transgenic mice expressing high levels of DDIT3, which lacks the FUS domain, do not develop any tumour but consistently show the accumulation of a glycoprotein material within the terminally differentiated adipocytes, a characteristic figure of liposarcomas associated with FUS-DDIT3. However, transgenic mice expressing the altered form of DDIT3-FUS (created by the in frame fusion of the FUS domain to the carboxy end of DDIT3) developed liposarcomas. The characteristics of the liposarcomas arising in the DDIT3-FUS mice were very similar to those previously observed in the FUS-DDIT3 transgenic mice indicating that the FUS domain is required not only for transformation but also influences the phenotype of the tumor cells. These results provide evidence that the FUS domain of FUS-DDIT3 plays a specific and critical role in the pathogenesis of liposarcoma. In this sense, when mice expressing the FUS domain are crossed with DDIT3-transgenic mice to generate the double-transgenic FUSxDDIT3, these animals develop liposarcoma. These results provide genetic evidence that FUS and DDIT3 domains function in trans for the mutual restoration of liposarcoma, and identify a new mechanism of tumour-associated fusion genes which might have impact beyond myxoid liposarcoma.
You might also like
- Human Tumor Suppressor p53 and DNA Viruses: RreeviiewDocument19 pagesHuman Tumor Suppressor p53 and DNA Viruses: RreeviiewEduardo LópezNo ratings yet
- Influence of FOX genes on aging and aging-associated diseasesFrom EverandInfluence of FOX genes on aging and aging-associated diseasesNo ratings yet
- Artículo 2Document11 pagesArtículo 2Manuel Andres Hernandez HurtadoNo ratings yet
- Pathology SWTDocument4 pagesPathology SWTkishorechandraNo ratings yet
- Critical Reviews in Oral Biology & Medicine: Matrix Metalloproteinases: A ReviewDocument55 pagesCritical Reviews in Oral Biology & Medicine: Matrix Metalloproteinases: A ReviewdrmacrohardNo ratings yet
- Funcionslidad de p53Document20 pagesFuncionslidad de p53macritoNo ratings yet
- DNA RepairDocument40 pagesDNA RepairANSH RAIZADA 20BOE10026No ratings yet
- LAMP GENE - Structure, Function and Involvement in CancerDocument5 pagesLAMP GENE - Structure, Function and Involvement in CancerIJRASETPublicationsNo ratings yet
- Foxp3 ThesisDocument7 pagesFoxp3 Thesisgja8e2sv100% (2)
- Acquired Capability: Evading ApoptosisDocument5 pagesAcquired Capability: Evading ApoptosisKaren Joy MariñasNo ratings yet
- Clinicopathologic and Molecular Features of Six Cases of Phosphaturic Mesenchymal TumorDocument9 pagesClinicopathologic and Molecular Features of Six Cases of Phosphaturic Mesenchymal TumorSOUMYA DEYNo ratings yet
- Diz 2Document6 pagesDiz 2Dulce TulaNo ratings yet
- Research ProposalDocument7 pagesResearch ProposalBitan BiswasNo ratings yet
- Molecular BiologyDocument44 pagesMolecular BiologyYasmin BalochNo ratings yet
- 2016 NGS Predictive of FIBROSISDocument12 pages2016 NGS Predictive of FIBROSISmaomaochongNo ratings yet
- Modification of Drosophila p53 by SUMO Modulates Its Transactivation and Pro-Apoptotic FunctionsDocument9 pagesModification of Drosophila p53 by SUMO Modulates Its Transactivation and Pro-Apoptotic FunctionsGustavo FelpeNo ratings yet
- Review Article: Osteosarcoma: Cells-of-Origin, Cancer Stem Cells, and Targeted TherapiesDocument14 pagesReview Article: Osteosarcoma: Cells-of-Origin, Cancer Stem Cells, and Targeted TherapiesJavier Ernesto GuavaNo ratings yet
- Position Effects and Genetic DiseaseDocument1 pagePosition Effects and Genetic DiseaseSharyproNo ratings yet
- MACROGNATHIADocument9 pagesMACROGNATHIAHapsari Nur PrimastutiNo ratings yet
- Fragile X Syndrome: Causes, Diagnosis, Mechanisms, and TherapeuticsDocument9 pagesFragile X Syndrome: Causes, Diagnosis, Mechanisms, and TherapeuticshanzelNo ratings yet
- 1 - Genetics Lec.1Document28 pages1 - Genetics Lec.1Ammar AlnajjarNo ratings yet
- High Prevalence of p53 Exon 4 Mutations in Soft Tissue SarcomaDocument11 pagesHigh Prevalence of p53 Exon 4 Mutations in Soft Tissue SarcomaLaura ChristinaNo ratings yet
- MeiosisDocument6 pagesMeiosiskarlosbigmaxNo ratings yet
- Hum. Mol. Genet. 1999 Raymond 989 96Document8 pagesHum. Mol. Genet. 1999 Raymond 989 96John MistryNo ratings yet
- Role of Rbm3 Gene in CancerDocument40 pagesRole of Rbm3 Gene in CancerAnkit AgarwalNo ratings yet
- MS 1-1 MS 1-3: Christine Sers Burkhard Scharm, Reinhold SCH FerDocument47 pagesMS 1-1 MS 1-3: Christine Sers Burkhard Scharm, Reinhold SCH FerindilaNo ratings yet
- Fgfr3 Signaling in Achondroplasia: A ReviewDocument5 pagesFgfr3 Signaling in Achondroplasia: A Reviewnurfitriana apriliaNo ratings yet
- Genes 13 00165 With CoverDocument12 pagesGenes 13 00165 With CoverMaria Garcia FernandezNo ratings yet
- Genetic Alterations in The Adenoma-Carcinoma Sequence: Kathleen R. ChoDocument5 pagesGenetic Alterations in The Adenoma-Carcinoma Sequence: Kathleen R. ChoSantiago AldayNo ratings yet
- FRM F022 - Billings - 3118Document7 pagesFRM F022 - Billings - 3118stungstungNo ratings yet
- Tugas DR KamalDocument7 pagesTugas DR KamalZarin SafanahNo ratings yet
- 2021 22363 AuthorDocument28 pages2021 22363 AuthorGeorge LaliotisNo ratings yet
- Multivalent Epigenetic Marks Confer Microenvironment Responsive Epigenetic Plasticity To Ovarian Cancer CellsDocument15 pagesMultivalent Epigenetic Marks Confer Microenvironment Responsive Epigenetic Plasticity To Ovarian Cancer CellsJyoti DeshmukhNo ratings yet
- Role of CRISPRCas9 in Genetic Manipulation of ROS1 and EGFR Genes Using Synthego PlatformDocument6 pagesRole of CRISPRCas9 in Genetic Manipulation of ROS1 and EGFR Genes Using Synthego PlatformInternational Journal of Innovative Science and Research TechnologyNo ratings yet
- Her2/Neu (Also Known As Erbb-2) Stands For "Human Epidermal Growth Factor Receptor 2"Document4 pagesHer2/Neu (Also Known As Erbb-2) Stands For "Human Epidermal Growth Factor Receptor 2"Bima AnantaNo ratings yet
- NBME 16 FlashreviewDocument27 pagesNBME 16 Flashreviewilse torresNo ratings yet
- Histone Deacetylases Transcriptional Control, and Cancer.Document16 pagesHistone Deacetylases Transcriptional Control, and Cancer.jose_gladiusNo ratings yet
- Multiple Lipomas Linked To An RB1 Gene Mutation in A Large Pedigree With Low Penetrance RetinoblastomaDocument5 pagesMultiple Lipomas Linked To An RB1 Gene Mutation in A Large Pedigree With Low Penetrance RetinoblastomaMaria Burke-ChaussonNo ratings yet
- Boumediene Bouzahzah - 2001 - 274 PDFDocument15 pagesBoumediene Bouzahzah - 2001 - 274 PDFarnipahlawaniNo ratings yet
- Main Genome Editing Tools An Overview of The Literature, Future Applications and Ethical QuestionsDocument15 pagesMain Genome Editing Tools An Overview of The Literature, Future Applications and Ethical QuestionsBilge Rana AkbolatNo ratings yet
- Genetic Alterations and DNA Repair in Human Carcinogenesis: Kathleen Dixon, Elizabeth KoprasDocument8 pagesGenetic Alterations and DNA Repair in Human Carcinogenesis: Kathleen Dixon, Elizabeth Kopras1205lorenaNo ratings yet
- Mosaicism For GNAS Methylation Defects AssociatedDocument11 pagesMosaicism For GNAS Methylation Defects Associatedradu nicolaeNo ratings yet
- Epigenetic and Cancer Therapy, 2019Document12 pagesEpigenetic and Cancer Therapy, 2019AbsjeyNo ratings yet
- Kes - Martin A. - Mutations in The Adrenoleukodystrophy Gene (1997) (10.1002 - (Sici) 1098-1004 (1997) 9 - 63.0.co - 2-5) - Libgen - LiDocument12 pagesKes - Martin A. - Mutations in The Adrenoleukodystrophy Gene (1997) (10.1002 - (Sici) 1098-1004 (1997) 9 - 63.0.co - 2-5) - Libgen - LiLoplopNo ratings yet
- p53 Enters The MicroRNA WorldDocument5 pagesp53 Enters The MicroRNA WorldKenanNo ratings yet
- C-FRC Hippo 2020 NatComDocument15 pagesC-FRC Hippo 2020 NatCompnom43582No ratings yet
- Dex3 and Survivin-2B: Two Novel Splice Variants of The ApoptosisDocument6 pagesDex3 and Survivin-2B: Two Novel Splice Variants of The ApoptosisNona NonicaaNo ratings yet
- Lecture #10. The Biology of Cancer, p53Document49 pagesLecture #10. The Biology of Cancer, p53cafemedNo ratings yet
- Bacchetta Et Al-2016-Annals of The New York Academy of SciencesDocument18 pagesBacchetta Et Al-2016-Annals of The New York Academy of SciencesjessicaNo ratings yet
- Clinical Development of Biomarker To Detect Oral Carcinoma in Relation To Genetic Polymorphism at MMP-9Document6 pagesClinical Development of Biomarker To Detect Oral Carcinoma in Relation To Genetic Polymorphism at MMP-9Ijsrnet EditorialNo ratings yet
- Arti IngleDocument6 pagesArti Ingleshadow darkNo ratings yet
- Duchenne Muscular Dystrophy: Marek Michalak, Michal OpasDocument5 pagesDuchenne Muscular Dystrophy: Marek Michalak, Michal OpasZurezki Yuana YafieNo ratings yet
- Protein Diversity 1Document4 pagesProtein Diversity 1Lalitha RajeshNo ratings yet
- Understanding Familial and Non-Familial Renal Cell CancerDocument10 pagesUnderstanding Familial and Non-Familial Renal Cell CancerVanroNo ratings yet
- Tumor Suppressor Gene & Proto-OncogeneDocument61 pagesTumor Suppressor Gene & Proto-OncogeneKartthik ShanmugamNo ratings yet
- !protocol (CFA) Induction of Epithelial-Mesenchymal Transition (EMT) and Gli1 Expression in Head and Neck Squamous Cell Carcinoma Spheroid CulturesDocument11 pages!protocol (CFA) Induction of Epithelial-Mesenchymal Transition (EMT) and Gli1 Expression in Head and Neck Squamous Cell Carcinoma Spheroid CulturesMohamed El-AgamyNo ratings yet
- Developmental Genetics and Pharmacogeneticsdoc4301Document8 pagesDevelopmental Genetics and Pharmacogeneticsdoc4301sayednourNo ratings yet
- 10 5772@54652 PDFDocument32 pages10 5772@54652 PDFhollNo ratings yet
- Tugas Bu AniDocument23 pagesTugas Bu AniYon-SyuhandaNo ratings yet
- HALOPERIDOLDocument3 pagesHALOPERIDOLSonny Dizon PareñasNo ratings yet
- Vitamins - Functions, Deficiency, Food SourcesDocument5 pagesVitamins - Functions, Deficiency, Food SourcesSonny Dizon PareñasNo ratings yet
- Mineral Function Deficiency Over Food Source MacromineralsDocument2 pagesMineral Function Deficiency Over Food Source MacromineralsSonny Dizon PareñasNo ratings yet
- Health Pattern Before Hospitalization During HospitalizationDocument1 pageHealth Pattern Before Hospitalization During HospitalizationSonny Dizon PareñasNo ratings yet
- Bleeding CholeDocument1 pageBleeding CholeSonny Dizon PareñasNo ratings yet
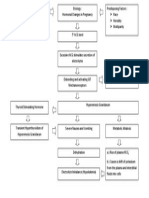
- Pathophysiology of Hyperemesis Gravidarum DiagramDocument1 pagePathophysiology of Hyperemesis Gravidarum DiagramSonny Dizon Pareñas100% (11)
- NCP 2013 ObDocument1 pageNCP 2013 ObSonny Dizon Pareñas100% (2)
- Chole NCPDocument20 pagesChole NCPSonny Dizon Pareñas100% (1)
- Macrominerals and Microminerals NotesDocument3 pagesMacrominerals and Microminerals NotesSonny Dizon PareñasNo ratings yet
- Pathophysiology of Hyperemesis Gravidarum DiagramDocument1 pagePathophysiology of Hyperemesis Gravidarum DiagramSonny Dizon Pareñas100% (11)
- Name Classification Action Indication Adverse Effect Contraindication Nursing ConsiderationDocument2 pagesName Classification Action Indication Adverse Effect Contraindication Nursing ConsiderationSonny Dizon PareñasNo ratings yet
- Assessment Nursing Diagnosis Inference Planning Intervention Rationale EvaluationDocument2 pagesAssessment Nursing Diagnosis Inference Planning Intervention Rationale EvaluationSonny Dizon PareñasNo ratings yet
- MalariaDocument1 pageMalariaSonny Dizon PareñasNo ratings yet
- Pelvic Organ ProlapseDocument9 pagesPelvic Organ ProlapseSonny Dizon PareñasNo ratings yet
- Name Classification Action Indication Adverse Effect Contraindication Nursing ConsiderationDocument3 pagesName Classification Action Indication Adverse Effect Contraindication Nursing ConsiderationSonny Dizon PareñasNo ratings yet
- Classification: by DegreeDocument10 pagesClassification: by DegreeSonny Dizon PareñasNo ratings yet
- Amyotrophic Lateral Sclerosis - Mechanisms and Therapeutics in The Epigenomic EraDocument14 pagesAmyotrophic Lateral Sclerosis - Mechanisms and Therapeutics in The Epigenomic EraEduardo Martinez VillalvazoNo ratings yet
- Report PDFDocument97 pagesReport PDFMark McKinsnkeyNo ratings yet
- ALSDocument14 pagesALSZé CunhaNo ratings yet
- Behavioral Variant Frontotemporal Dementia.7Document25 pagesBehavioral Variant Frontotemporal Dementia.7wmtxbbNo ratings yet
- Alzheimer Disease Sourcebook 5th Ed. - A. Sutton (Omnigraphics, 2011) BBSDocument335 pagesAlzheimer Disease Sourcebook 5th Ed. - A. Sutton (Omnigraphics, 2011) BBSvkarmoNo ratings yet
- Frontotemporal DementiaDocument11 pagesFrontotemporal DementiaLudovico MineoNo ratings yet
- Myxoid LiposarcomaDocument4 pagesMyxoid LiposarcomaSonny Dizon PareñasNo ratings yet
- Dementia FrontotemporalDocument24 pagesDementia FrontotemporalTeologia GamalielNo ratings yet
- Alzheimer Disease Including Focal Presentations PDFDocument14 pagesAlzheimer Disease Including Focal Presentations PDFpiedrahitasaNo ratings yet
- Drosophila Melanogaster Models of Motor Neuron Disease 2013 (Edited by Ruben Cauchi)Document239 pagesDrosophila Melanogaster Models of Motor Neuron Disease 2013 (Edited by Ruben Cauchi)Dr.Gill_Grissom100% (1)
- COVID-19 RNA-Based Vaccines and The Risk of Prion Disease 1503Document3 pagesCOVID-19 RNA-Based Vaccines and The Risk of Prion Disease 1503jermNo ratings yet
- Kuzma-Kozakiewicz (2011) New Therapeutic Targets For ALS PDFDocument17 pagesKuzma-Kozakiewicz (2011) New Therapeutic Targets For ALS PDFLyly MagnanNo ratings yet
- Sarcoma de Ewing S NEJMDocument11 pagesSarcoma de Ewing S NEJMbanzethyskyNo ratings yet
- TDP-43 and FUS in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia PDFDocument13 pagesTDP-43 and FUS in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia PDFmiliindianuNo ratings yet
- Return of the God Hypothesis: Three Scientific Discoveries That Reveal the Mind Behind the UniverseFrom EverandReturn of the God Hypothesis: Three Scientific Discoveries That Reveal the Mind Behind the UniverseRating: 4.5 out of 5 stars4.5/5 (52)
- The Rise and Fall of the Dinosaurs: A New History of a Lost WorldFrom EverandThe Rise and Fall of the Dinosaurs: A New History of a Lost WorldRating: 4 out of 5 stars4/5 (597)
- Why We Die: The New Science of Aging and the Quest for ImmortalityFrom EverandWhy We Die: The New Science of Aging and the Quest for ImmortalityRating: 4 out of 5 stars4/5 (5)
- A Brief History of Intelligence: Evolution, AI, and the Five Breakthroughs That Made Our BrainsFrom EverandA Brief History of Intelligence: Evolution, AI, and the Five Breakthroughs That Made Our BrainsRating: 4.5 out of 5 stars4.5/5 (6)
- When the Body Says No by Gabor Maté: Key Takeaways, Summary & AnalysisFrom EverandWhen the Body Says No by Gabor Maté: Key Takeaways, Summary & AnalysisRating: 3.5 out of 5 stars3.5/5 (2)
- Gut: the new and revised Sunday Times bestsellerFrom EverandGut: the new and revised Sunday Times bestsellerRating: 4 out of 5 stars4/5 (393)
- Buddha's Brain: The Practical Neuroscience of Happiness, Love & WisdomFrom EverandBuddha's Brain: The Practical Neuroscience of Happiness, Love & WisdomRating: 4 out of 5 stars4/5 (216)
- 10% Human: How Your Body's Microbes Hold the Key to Health and HappinessFrom Everand10% Human: How Your Body's Microbes Hold the Key to Health and HappinessRating: 4 out of 5 stars4/5 (33)
- The Molecule of More: How a Single Chemical in Your Brain Drives Love, Sex, and Creativity--and Will Determine the Fate of the Human RaceFrom EverandThe Molecule of More: How a Single Chemical in Your Brain Drives Love, Sex, and Creativity--and Will Determine the Fate of the Human RaceRating: 4.5 out of 5 stars4.5/5 (517)
- Gut: The Inside Story of Our Body's Most Underrated Organ (Revised Edition)From EverandGut: The Inside Story of Our Body's Most Underrated Organ (Revised Edition)Rating: 4 out of 5 stars4/5 (411)
- Tales from Both Sides of the Brain: A Life in NeuroscienceFrom EverandTales from Both Sides of the Brain: A Life in NeuroscienceRating: 3 out of 5 stars3/5 (18)
- All That Remains: A Renowned Forensic Scientist on Death, Mortality, and Solving CrimesFrom EverandAll That Remains: A Renowned Forensic Scientist on Death, Mortality, and Solving CrimesRating: 4.5 out of 5 stars4.5/5 (397)
- Undeniable: How Biology Confirms Our Intuition That Life Is DesignedFrom EverandUndeniable: How Biology Confirms Our Intuition That Life Is DesignedRating: 4 out of 5 stars4/5 (11)
- Seven and a Half Lessons About the BrainFrom EverandSeven and a Half Lessons About the BrainRating: 4 out of 5 stars4/5 (111)
- The Ancestor's Tale: A Pilgrimage to the Dawn of EvolutionFrom EverandThe Ancestor's Tale: A Pilgrimage to the Dawn of EvolutionRating: 4 out of 5 stars4/5 (812)
- Who's in Charge?: Free Will and the Science of the BrainFrom EverandWho's in Charge?: Free Will and the Science of the BrainRating: 4 out of 5 stars4/5 (65)
- The Other Side of Normal: How Biology Is Providing the Clues to Unlock the Secrets of Normal and Abnormal BehaviorFrom EverandThe Other Side of Normal: How Biology Is Providing the Clues to Unlock the Secrets of Normal and Abnormal BehaviorNo ratings yet
- Lymph & Longevity: The Untapped Secret to HealthFrom EverandLymph & Longevity: The Untapped Secret to HealthRating: 4.5 out of 5 stars4.5/5 (13)
- The Lives of Bees: The Untold Story of the Honey Bee in the WildFrom EverandThe Lives of Bees: The Untold Story of the Honey Bee in the WildRating: 4.5 out of 5 stars4.5/5 (44)
- The Consciousness Instinct: Unraveling the Mystery of How the Brain Makes the MindFrom EverandThe Consciousness Instinct: Unraveling the Mystery of How the Brain Makes the MindRating: 4.5 out of 5 stars4.5/5 (93)
- Moral Tribes: Emotion, Reason, and the Gap Between Us and ThemFrom EverandMoral Tribes: Emotion, Reason, and the Gap Between Us and ThemRating: 4.5 out of 5 stars4.5/5 (115)
- Why We Sleep: Unlocking the Power of Sleep and DreamsFrom EverandWhy We Sleep: Unlocking the Power of Sleep and DreamsRating: 4.5 out of 5 stars4.5/5 (2083)
- Good Without God: What a Billion Nonreligious People Do BelieveFrom EverandGood Without God: What a Billion Nonreligious People Do BelieveRating: 4 out of 5 stars4/5 (66)