Professional Documents
Culture Documents
Biophysics Lecture2
Uploaded by
Sarthak SubhankarCopyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Biophysics Lecture2
Uploaded by
Sarthak SubhankarCopyright:
Available Formats
24 MACROMOLECULES
2.1.2 The Gaussian Chain Model
Important Concepts: uctuating bond, Hamiltonian for the
Gaussian Chain Model
We consider a chain made up of orientationally uncorrelated (freely-jointed) links
where the length of any link vector is no longer constant but has a probability
distribution
G(r) =
_
3
2b
2
_
3/2
exp
_
3r
2
2b
2
_
(2.36)
with the expectation for the link length being
< r
2
>= b
2
. (2.37)
The probability distribution for the end-to-end vector is then
P(R
e
) = P({r
n
}) (2.38)
=
N
n=1
_
3
2b
2
_
3/2
exp
_
3r
2
n
2b
2
_
(2.39)
=
_
3
2b
2
_
3/2
exp
_
n=1
3(R
n
R
n1
)
2
2b
2
_
(2.40)
and hence for the entropy
S = lnP =
N
n=1
ln P(r
n
) (2.41)
= const
3
2b
2
N
n=1
r
2
n
. (2.42)
From this we obtain the free energy
F({r
n
}) = E +
3T
2b
2
N
n=1
r
2
n
(2.43)
with the internal energy E being independent of {r
n
}.
Prof. Heermann, Heidelberg University
2.1 GENERAL PROPERTIES OF MACROMOLECULES 25
Hence we obtain the same equilibrium distribution as for the freely-jointed chain.
Eq (2.40) also results if we start off with the Hamiltonian for a chain of springs
H =
3
2
k
B
T
b
2
N
n=1
(R
n
R
n1
)
2
(2.44)
and we also obtain the scaling of the end-to-end distance
< R
2
e
> N . (2.45)
Further Reading 2.1.4 (Continuous scales)
For the later development we note the generalization to all scales for the Gaussian
model at xed contour length L
P(R) =
_
r(L)=R
r(0)=0
D[r(s)] exp
_
3L
2b
2
_
L
0
ds
_
r(s)
s
_
2
_
, (2.46)
where r(s) denotes the contour or conformation of the chain. This is solved by
P(R) =
_
3
2b
2
_
3/2
exp
_
3R
2
2b
2
_
(2.47)
and is the proper continuum limit of the FJC, where the limits N and b 0
were taken, such that Nb
2
stayed nite.
We further generalize the effective Hamiltonian
H =
3
2
k
B
T
b
2
_
L
0
ds r
2
(s) (2.48)
where the derivation is to be taken along the contour.
2.1.3 Worm-like Chain Model
Important Concepts: bending rigidity and the relation with the
persistence length
A short-coming of the above models (besides they being phantom chains, i.e.
no self-avoidance) is that there is no intrinsic stiffness. Intuitively, we expect a
bending of the chain to cost energy. A model that provides this is the worm-like
chain model (WLC)
Prof. Heermann, Heidelberg University
26 MACROMOLECULES
R(s)
r(s)
r
n
r
n+1
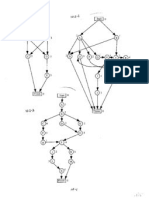
Figure 2.10: Denition of the coordinates used to describe the chain as a con-
tinuous curve
H =
N1
n=1
r
n
r
n+1
(2.49)
which is simply the one-dimensional Heisenberg model for ferromagnets. Here
|r
n
| = b. This model can be treated in the continuum limit where N , b 0
and with
/N = constant , (2.50)
keeping the contour length also constant. Using
r
n+1
r
n
=
1
2
[(r
n
r
n+1
)
2
2b
2
] (2.51)
we have
H = lim
b0,,N
b
2
N1
n=1
b
_
r
n
r
n+1
b
_
2
. (2.52)
To cross over to the continuum limit we use the tangent vector with the arc length
s
r(s)
s
= lim
b0
_
r
n+1
r
n
b
_
(2.53)
Prof. Heermann, Heidelberg University
2.1 GENERAL PROPERTIES OF MACROMOLECULES 27
and
N1
n=1
b
_
L
0
ds to nd
H =
2
_
L
0
ds
_
r(s)
s
_
2
=
2
_
L
0
ds
_
2
R(s)
s
2
_
2
(2.54)
with the bending modulus = b.
Thus the partition function is given by
Z =
_
D[r(s)](|r(s)| 1)exp(H[r(s)]) . (2.55)
The bending modulus must have a relation with the persistence length. To nd
this relation we need to calculate the correlation function
< r(s)r(s
) > exp(|s s
|/
p
) . (2.56)
We can nowcalculate the mean squared end-to-end-distance and the mean squared
radius of gyration
< R
2
e
> = <
__
L
0
dsr(s)
_
2
> (2.57)
=
_
L
0
ds
_
L
0
ds
< r(s) r(s
) > (2.58)
= 2
2
p
_
L
p
1 + e
L/p
_
(2.59)
= L
2
f
D
_
L
p
_
, (2.60)
where f
D
(x) = 2(x 1 + e
x
)/x
2
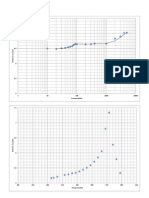
being the Debye-function (see gure 2.11).
Further Reading 2.1.5 (Lattice Chains)
We can also go about calculating the average end-to-end distance staying discrete
(Kratky-Porod model [36] ). For this we write
H =
N1
n=1
r
n
r
n+1
=
N
n=1
cos
n
. (2.61)
The partition function is given by
Z
N
=
N
n=1
_
d
n
e
cos n
= Q
N
l
(2.62)
Prof. Heermann, Heidelberg University
28 MACROMOLECULES
30 10
x
60 40 50
fd(x)
1
0,8
0,6
20
0,4
0,2
0
TheDebyefunction
Figure 2.11: The Debye-function
with
Q
l
= 2
_
1
1
d(cos)e
cos
= 4
sinh
. (2.63)
With this we can calculate the distance
< R
2
e
>= b
2
< (
r
n
)
2
>= b
2
n,m
< r
n
r
m
> (2.64)
again as the correlation between the directions of bonds. For the nearest neighbour
correlation we have
< r
n
r
n+1
> = < cos > (2.65)
=
_
dcos e
cos
_
de
cos
(2.66)
=
d lnQ
l
d()
(2.67)
= coth() 1/ (2.68)
= f
L
() , (2.69)
where f
L
is the Langevin function. In general the correlation is given by [37]
Prof. Heermann, Heidelberg University
2.1 GENERAL PROPERTIES OF MACROMOLECULES 29
< r
n
r
n+1
>= e
N ln(1/c)
, (2.70)
which can be written in terms of the persistence length
p
< r
n
r
n+1
>= e
Nb/p
(2.71)
with
p
=
b
ln(1/c)
(2.72)
If the macromolecule is stiff >> 1 and
c 1 k
B
T/ , (2.73)
then
ln(1/c) k
B
T/ (2.74)
and thus
p
= b . (2.75)
Let us rewrite the end-to-end distance in terms of c
1
b
2
< R
2
e
>=
N
i=1
_
i1
j=1
c
ij
+ 1 +
N
j=i+1
c
ji
_
, (2.76)
then for most i the two sums in the braces can be replaced by
k=1
=
c
1c
and
with this we obtain
< R
2
e
> Nb
2
_
1 +
2c
1 c
_
= Nb
2
1 + c
1 c
. (2.77)
1+c
1c
is the Flory factor. The Kuhn segment length l
K
is then given by
l
K
=
1 + c
1 c
b . (2.78)
Prof. Heermann, Heidelberg University
30 MACROMOLECULES
P
b
t
n
b
t
n
P`
Figure 2.12: Motion of the Fernet triad along a curve
Since we are dealing in some sense with mathematical curves in three dimen-
sional space recall that a curve in space is mathematically determined by the two
parameters: curvature and torsion ( the basic triad {t
k
})
db
ds
= n ,
dn
ds
= t + b ,
t
ds
= n (2.79)
Choosing = = 0 we obtain a straight line, r = 1/ and = 0 for a circle
with radius r.
Consider the case of a helix (see gure 2.13) with
r(s) =
_
r cos
_
qs
_
(qr)
2
+ 1
_
, r sin
_
qs
_
(qr)
2
+ 1
_
,
s
_
(qr)
2
+ 1
_
(2.80)
and
(s) =
rq
2
1 + (qr)
2
(2.81)
(s) =
q
1 + (qr)
2
(2.82)
Clearly as r 0 we must approach a straight line. The curvature vanishes indeed
in this limit but the torsion does not.
Prof. Heermann, Heidelberg University
2.1 GENERAL PROPERTIES OF MACROMOLECULES 31
Pitch p
Radius r
Figure 2.13: Denition of the parameters for a helix
We can generalize the Fernet-frame introducing a third parameter
dt
i
(s)
ds
=
j,k
ijk
j
(s)t
k
(s) (2.83)
where
1
= cos ,
2
= sin ,
3
= +
d
ds
(2.84)
This could be used to dene an elastic energy with spontaneous curvature and
torsion {
0k
} (the deviation from equilibrium is denoted by
k
=
k
0k
U =
1
2
k
b
k
_
L
0
2
k
ds (2.85)
The parameters b
k
dene the rigidity. Dening
Prof. Heermann, Heidelberg University
32 MACROMOLECULES
A =
_
_
0
0
0
_
_
(2.86)
with
A
ij
=
ijk
k
(2.87)
we can develop also a numerical procedure to generate curves that have a pre-
scribed spontaneous curvature with uctuations. For this let v
x
= (t
x
1
, t
x
2
, t
x
3
, ) etc.
then we write the equation 2.83 as
dv
i
ds
= Av
i
(2.88)
which can be discretized as
v
i
(s + ds) = Ov
i
(s) (2.89)
with
O = (1 +
ds
2
A)(1
ds
2
A)
1
(2.90)
Note that O is an orthogonal matrix guaranteeing that we consistently generate
orthogonal triads.
In gure is depicted the case for a curve where the spontaneous curvature is set
such that a circle is preferred.
2.1.4 Self-Avoiding Chains
The above models all lack the excluded volume interaction between the monomers.
Indeed, the ubiquitous polymer model is a self-avoiding random walk.
Let U(R
n
R
m
) be a monomer-monomer interaction potential assumed to handle
the excluded volume. We treat the polymer as gas of disconnected monomers
conned within the same volume V as the polymer coil. If
R denotes the average
extent of the chain then the chain occupies a volume V =
R
3
. Let F
c
be the term
that arises from the chemical work done by initially preparing the monomers in
Prof. Heermann, Heidelberg University
You might also like
- Operator SplittingDocument109 pagesOperator SplittingSarthak SubhankarNo ratings yet
- Lecture 05Document6 pagesLecture 05Sarthak SubhankarNo ratings yet
- Lesson 4 Extinction & ScatteringDocument43 pagesLesson 4 Extinction & ScatteringSarthak SubhankarNo ratings yet
- Experiment 1Document4 pagesExperiment 1Sarthak SubhankarNo ratings yet
- Document 1experimentDocument2 pagesDocument 1experimentSarthak SubhankarNo ratings yet
- Web Book On LightDocument345 pagesWeb Book On LightSarthak SubhankarNo ratings yet
- OR Ch10Document38 pagesOR Ch10McClelland KempNo ratings yet
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceFrom EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceRating: 4 out of 5 stars4/5 (895)
- Never Split the Difference: Negotiating As If Your Life Depended On ItFrom EverandNever Split the Difference: Negotiating As If Your Life Depended On ItRating: 4.5 out of 5 stars4.5/5 (838)
- The Yellow House: A Memoir (2019 National Book Award Winner)From EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Rating: 4 out of 5 stars4/5 (98)
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeFrom EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeRating: 4 out of 5 stars4/5 (5794)
- Shoe Dog: A Memoir by the Creator of NikeFrom EverandShoe Dog: A Memoir by the Creator of NikeRating: 4.5 out of 5 stars4.5/5 (537)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaFrom EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaRating: 4.5 out of 5 stars4.5/5 (266)
- The Little Book of Hygge: Danish Secrets to Happy LivingFrom EverandThe Little Book of Hygge: Danish Secrets to Happy LivingRating: 3.5 out of 5 stars3.5/5 (400)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureFrom EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureRating: 4.5 out of 5 stars4.5/5 (474)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryFrom EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryRating: 3.5 out of 5 stars3.5/5 (231)
- Grit: The Power of Passion and PerseveranceFrom EverandGrit: The Power of Passion and PerseveranceRating: 4 out of 5 stars4/5 (588)
- The Emperor of All Maladies: A Biography of CancerFrom EverandThe Emperor of All Maladies: A Biography of CancerRating: 4.5 out of 5 stars4.5/5 (271)
- The Unwinding: An Inner History of the New AmericaFrom EverandThe Unwinding: An Inner History of the New AmericaRating: 4 out of 5 stars4/5 (45)
- On Fire: The (Burning) Case for a Green New DealFrom EverandOn Fire: The (Burning) Case for a Green New DealRating: 4 out of 5 stars4/5 (74)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersFrom EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersRating: 4.5 out of 5 stars4.5/5 (344)
- Team of Rivals: The Political Genius of Abraham LincolnFrom EverandTeam of Rivals: The Political Genius of Abraham LincolnRating: 4.5 out of 5 stars4.5/5 (234)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreFrom EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreRating: 4 out of 5 stars4/5 (1090)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyFrom EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyRating: 3.5 out of 5 stars3.5/5 (2259)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)From EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Rating: 4.5 out of 5 stars4.5/5 (121)
- Her Body and Other Parties: StoriesFrom EverandHer Body and Other Parties: StoriesRating: 4 out of 5 stars4/5 (821)
- Chapter 4 - Modes of ExtinguishmentDocument19 pagesChapter 4 - Modes of ExtinguishmentcartyeolNo ratings yet
- Martin Luther King. Answer KeyDocument1 pageMartin Luther King. Answer KeyOleg MuhendisNo ratings yet
- Anthropometry in DesignDocument45 pagesAnthropometry in DesignShalini YadavNo ratings yet
- Customer Communication.Document3 pagesCustomer Communication.wilson wambuguNo ratings yet
- Sorrows of A Trophy WifeDocument786 pagesSorrows of A Trophy WifeAngel MilanNo ratings yet
- Upholding Malay Language Strengthening Command English LanguageDocument12 pagesUpholding Malay Language Strengthening Command English LanguagemucheliaNo ratings yet
- Watchtower: What Say The Scriptures About Spiritualism - Proofs That Its Demonism - 1897Document126 pagesWatchtower: What Say The Scriptures About Spiritualism - Proofs That Its Demonism - 1897sirjsslutNo ratings yet
- DPP-Control and Coordination (Prashant Kirad)Document9 pagesDPP-Control and Coordination (Prashant Kirad)ABHI TECH 983No ratings yet
- I Semester Financial Accounting - Bpz1A: Theory 15 Problems 60Document28 pagesI Semester Financial Accounting - Bpz1A: Theory 15 Problems 60Blaze MysticNo ratings yet
- Oracle Database Performance Tuning Advanced Features and Best Practices For DbasDocument303 pagesOracle Database Performance Tuning Advanced Features and Best Practices For Dbassss pppNo ratings yet
- Phed 239 Syllabus s14-1st Half-3Document7 pagesPhed 239 Syllabus s14-1st Half-3api-249627241No ratings yet
- Quantitative Option Strategies: Marco Avellaneda G63.2936.001 Spring Semester 2009Document29 pagesQuantitative Option Strategies: Marco Avellaneda G63.2936.001 Spring Semester 2009Adi MNo ratings yet
- 11 Rabino v. Cruz 222 SCRA 493Document4 pages11 Rabino v. Cruz 222 SCRA 493Joshua Janine LugtuNo ratings yet
- Self Incompatibility: Dr. L.K.GangwarDocument38 pagesSelf Incompatibility: Dr. L.K.GangwarSiddhant Singh100% (1)
- Catalog Encoder MILEDocument1 pageCatalog Encoder MILELucas SuplinoNo ratings yet
- John Zink - Flare - Upstream - ProductionDocument20 pagesJohn Zink - Flare - Upstream - ProductionJose Bijoy100% (2)
- In The HeightsDocument15 pagesIn The Heightsapi-548206714No ratings yet
- Cisco SF300Document401 pagesCisco SF300Rodney PolingNo ratings yet
- Tutorial MATLAB Matrix NotationDocument10 pagesTutorial MATLAB Matrix NotationgeorgesmaccarioNo ratings yet
- Module 5 Network Design in The Supply ChainDocument15 pagesModule 5 Network Design in The Supply ChainstraheelNo ratings yet
- Rocketology ProjectDocument6 pagesRocketology ProjectJosue Grana0% (1)
- Survey of Dealers and Subdealers of Kajaria Vitrified Tiles To Know The Market Trend and Potential in Ghaziabad and NoidaDocument13 pagesSurvey of Dealers and Subdealers of Kajaria Vitrified Tiles To Know The Market Trend and Potential in Ghaziabad and Noidajdjeet4100% (1)
- 30 Feet: High Elf MediumDocument10 pages30 Feet: High Elf MediumAli Kaan (Uyau)No ratings yet
- Ethical Hacking and Penetration Testing Guide - Baloch, RafayDocument2 pagesEthical Hacking and Penetration Testing Guide - Baloch, RafayAyush PoptaniNo ratings yet
- Ieo Sample Paper Class-10Document2 pagesIeo Sample Paper Class-10jairusr100% (1)
- Cara Urutan Minyak LintahDocument4 pagesCara Urutan Minyak Lintahaerohel100% (1)
- Visual Development Milestones and Visual Acuity Assessment in ChildrenDocument2 pagesVisual Development Milestones and Visual Acuity Assessment in ChildrenNikhil Maha DevanNo ratings yet
- Social Defence Vision 2020: Dr. Hira Singh Former Director National Institute of Social DefenceDocument19 pagesSocial Defence Vision 2020: Dr. Hira Singh Former Director National Institute of Social DefencePASION Jovelyn M.No ratings yet
- Pedagogue in The ArchiveDocument42 pagesPedagogue in The ArchivePaula LombardiNo ratings yet
- Culture Shock Refers To The Feelings of Confusion and Uncertainty That You Experience When You Come Into Contact With A New CultureDocument13 pagesCulture Shock Refers To The Feelings of Confusion and Uncertainty That You Experience When You Come Into Contact With A New CultureRetno WulanNo ratings yet