Professional Documents
Culture Documents
s516 14s-Homework-21
Uploaded by
api-253411445Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
s516 14s-Homework-21
Uploaded by
api-253411445Copyright:
Available Formats
Homework 2: Nonlinear Regression & Residuals
Dr. Timothy R. Johnson Spring Semester, 2014
This homework assignment is due no later than 3:00 on Wednesday, March 5th. Please read the instructions below carefully.
Homework Instructions
This homework assignment is due no later than 3:00 on Wednesday, March 5th. Late assignments will only be accepted in extreme circumstances and only if arrangements have been made in advance. Your solutions must be typed and very neatly organized. I will not try to infer your solutions if it they are not clearly presented. Equations need not be typeset perfectly but they should be clear. You may substitute letters for symbols (e.g., b1 for 1 ), and you may write-out equations (neatly) by hand if necessary. Include with your solutions you must include the relevant R output and the R scripts that created them. Include these within the text of your solutions using cut-and-paste. Try to include only the relevant output. Use a monospace font (e.g., Consolas or Courier) for R scripts and output for clarity, but only for R scripts and output. It is permitted for you to discuss the homework with other students in the course. However you must still write your own R scripts, produce your own output, and write up your own solutions. You are welcome to ask me questions concerning the homework. I will be particularly open to helping with any R problems. I want to evaluate your understanding of applied regression, not R, but part of the purpose of the homework assignments is to get you to exercise using R. If you email me with a R question, it may be helpful for you to include with your email your full R script so that I can replicate your problem. The Statistics Assistance Center (SAC) and Statistical Consulting Center (SCC) are not designed to accommodate this course. Direct all questions to me.
homework 2 : nonlinear regression & residuals
Power-Transformations via Nonlinear Regression
For this problem you will be revisiting the ToothGrowth data frame.1 Consider the following model:
> model <- lm(len ~ supp + dose + supp:dose, data = ToothGrowth) > summary(model) Estimate Std. Error t value Pr(>|t|) (Intercept) suppVC dose suppVC:dose 11.550 -8.255 7.811 3.904 1.581 2.236 1.195 1.691 7.304 1.09e-09 *** -3.691 0.000507 *** 6.534 2.03e-08 *** 2.309 0.024631 *
Load it with data(ToothGrowth). It is part of the base R distribution so you do not need to load a package.
1
From the output it can be seen that the model is + + ( + )dose , if supp = VC, 0 2 3 1 i i E(lengthi ) = 0 + 2 dosei , if suppi = OJ. In the following problems you will be using the nls function in R to estimate this model and some extensions of it. 1. Use nls to estimate the model above. Do this using the ifelse function rather than constructing indicator variables manually. Conrm you have specied your model correctly by comparing the summary of your nls object to that of the lm object above. 2. Recall that we looked at this model and concluded that a linear relationship between expected length and dose may not be appropriate.2 One method of dealing with a nonlinear relationship is to transform an explanatory variable to linearize the relationship. In the exercise you considered the model + + ( + ) g(dose ), if supp = VC, 0 2 3 1 i i E(lengthi ) = 0 + 2 g(dosei ), if supp = OJ,
i
Note in the plot from the in-class exercise where we delt with these data and this model that when dose is 1mg that the model appears to underestimate expected length.
2
where g( x ) = log2 ( x ). This time you will use the function g( x ) = log( x ).3 R knows this function simply as log so this transformation can be incorporated into the model as, for example,
> model <- lm(len ~ supp + log(dose) + supp:log(dose), data = ToothGrowth)
Use nls to estimate the model above similarly to how you used nls in the previous problem. Report the results from summary for the lm and nls objects to conrm that you specied your model correctly.
The function log is the natural log with base e which is also written sometimes as loge or ln. It can be shown that log2 ( x ) = loge ( x )/ loge (2). Because logarithms with different bases are proportional we can usually use whatever base is convenient in a linear regression model since a change in base will change the scale of some of the j parameters analogous to changing, say, meters to centimeters but will not change the model overall in the sense that the predicted values will be the same.
3
homework 2 : nonlinear regression & residuals
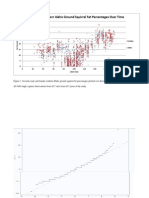
3. Consider the following transformation function. log( x ), if = 0, g( x, ) = ( x 1)/, if = 0. The quantity is a parameter that denes the transformation.4 Some simple functions are special cases of this function such as g( x, 1) = 1/y, g( x, 0) = log( x ), g( x, 0.5) = x, and g( x, 1) = y 1.5 Figure 1 illustrates the effect of the transformation for several values of . R does not know this function, but you can teach it by running the following in your script.6
g <- function(x, lambda) { if (lambda == 0) { x <- log(x) } else { x <- (x^lambda - 1)/lambda } return(x) }
In case you are curious, the unusual case-wise nature of the function is because we cannot divide by zero, but it can be shown that
4
lim
x 1 = log( x ),
so we dene g( x, ) = log( x ) if = 0.
5 When = 1 there is no transformation of the explanatory variable. For example,
0 + 1 g ( x i , 1) = 0 + 1 ( x i 1)
= 0 + 1 xi
where 0 = 0 1 . It simply result in an inconsequential change in the parameterization of the model.
1.5
1 0.5 0 0 .5 1 2
g (x, )
1.5 1.0 0.5
0.0
0.5
1.0
Now you could do something like
> model <- lm(len ~ supp + g(dose,0) + supp:g(dose,0), data = ToothGrowth)
0.5
1.0
1.5
2.0
x
Figure 1: Illustration of transformation function g for different values of . R is a highly developed programming language as well as a statistical computing environment.
6
if you wanted to use the log transformation for dose. Now if is specied then the model is linear. However the model is nonlinear if is to be estimated along with 0 , 1 , 2 , and 3 . Using your R code from the previous problem as a template, use nls to estimate 0 , 1 , 2 , 3 , and . Report the summary for the nls object.7 4. Using the script for the Puromycin example from the in-class exercise as a template, create a gure that shows the raw data and the predicted values from the nonlinear model you used in the previous problem.8 5. Consider the null hypothesis H0 : 3 = 0. Rejecting this null hypothesis supports the observation that expected tooth length does not increase at the same rate with dose when comparing the two supplement types. As we discussed in class, the test given by summary is not reliable for small n. Use the following two methods to test the null hypothesis above: (a) a condence interval, (b) the anova with an appropriate full and null model. Summarize the results of both tests assuming = 0.05.
For starting values, I would recommend using the linear model with a xed value of (e.g., = 1 or = 0), and then using the estimates from that analysis for starting values for the estimates of 0 , 1 , 2 , and 3 in your nonlinear model. For the starting value of use the same value you used in your linear model.
7
To save your plot select Copy Plot to Clipboard... or Save Plot as Image... option from the Export menu above the plot in RStudio. The formats for the latter depends on what your word processor can import. JPEG and TIFF are usually doable. Metale, when it works, is usually higher quality. For publication-quality graphics I usually use EPS or PDF. You may nd that you will need to tinker with the width and height to get the appropriate scale.
8
homework 2 : nonlinear regression & residuals
Aerial Snow Geese Counting
The following problems concern data from an unpublished report for the Canadian Wildlife Service. A description of the study follows.
Aerial survey methods are regularly used to estimated the number of snow geese in their summer range areas west of Hudson Bay in Canada. To obtain estimates, small aircraft y over the range and, when a ock of geese is spotted, an experienced person estimates the number of geese in the ock. To investigate the reliability of this method of counting, an experiment was conducted in which an airplane carrying two observers ew over 45 ocks, and each observer made an independent estimate of the number of birds in each ock. Also, a photograph of the ock was taken so that an exact count of the number of birds in the ock could be made.
The goal will be to model the expected observer count as a function of the true count (i.e., the photo count) and the observer (i.e., observer 1 or 2).9 I have prepared a R script le that will format the data, t a linear regression model, and plot the raw data, and the estimated model. It can be obtained at the following link.10
https://dl.dropboxusercontent.com/u/10884844/Homework/snowgeese.R
This sort of analysis, sometimes called calibration in the context of regression, can be used to evaluate the effectiveness of a cheaper/easier but less accurate measurement method (e.g., human estimated count) versus a more expensive/difcult but more accurate method (e.g., photo count).
9
Use this script, with necessary modications, to answer the following questions. For any problem that involves statistical graphics, include the gure(s) in your answers. 1. Examine a plot of the residuals against the predicted values. Does there appear to be any evidence of issues with either the validity of the model in terms of the relationship between the expected observer count and the true count, or in terms of the homoscedasticity assumption?11 Explain why or why not. When looking at the individual studentized residuals, do any observations look questionable leading you to check the source of the data to see if there might be, say, transcription errors? Finally produce a q-q plot of the studentized residuals to visually check the normality assumption. Is the q-q plot consistent with the assumption that the errors are (approximately) normally distributed? Why or why not? 2. Try using weighted least squares to resolve the apparent heteroscedasticity. You can specify the weights by using an option in lm such as
> model <- lm(y ~ x, data = mydata, weights = myweights)
You may need to install the alr3 and reshape2 packages. The reshape2 package contains the melt function which is very handy for reshaping data from short to long form. This is frequently useful for preparing data for regression modeling.
10
An easy way to make your plot would be to simply modify the x and y aesthetic parameters used by ggplot in the original R script.
11
where myweights is the name of the variable in the data frame you will use as weights.12 For the snow geese data, the variability appears to increase with the photo count. Perhaps a reasonable
Many other R functions for regression have an optional weight argument, although these do not necessarily result in weighted least squares in all cases. Weights have several other uses.
12
homework 2 : nonlinear regression & residuals
solution would be to assume that the variance is proportional to the photo count and to thus specify the weights as wi = 1 , photoi
where wi is the weight for the i-th observation and photoi is the photo count for the i-th observation. Try using these weights and inspect the plot of the studentized residuals to see if there is any improvement13 Comment on the effect of using the weights on the residual plot. 3. Another approach to dealing with heteroscedasticity is by transforming the response variable. Since the response variable (observer count) and the quantitative explanatory variable (photo count) are on the same scale, however, it might be reasonable to transform both variables in the same way. A couple of transformations that might work are the log and square root transformation. Transform both variables using something like the following.
> snowgeese.long$fcount <- f(snowgeese.long$count) > snowgeese.long$fphoto <- f(snowgeese.long$photo)
Standardized/studentized residuals account or the weights in the process of standardization so they can be used to determine if weighting helps resolve heteroscedasticity. This is not true for raw residuals.
13
where f is your chosen transformation (log is just log and square root is sqrt in R). Examine the plots of the studentized residuals against the predicted values and the q-q plots for each transformation and comment on if there appears to be any improvement in comparison to the plots you produced for the original data.14
Do not use weighted least squares for this problem. Use ordinary least squares.
14
You might also like
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeFrom EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeRating: 4 out of 5 stars4/5 (5794)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreFrom EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreRating: 4 out of 5 stars4/5 (1090)
- Never Split the Difference: Negotiating As If Your Life Depended On ItFrom EverandNever Split the Difference: Negotiating As If Your Life Depended On ItRating: 4.5 out of 5 stars4.5/5 (838)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceFrom EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceRating: 4 out of 5 stars4/5 (895)
- Grit: The Power of Passion and PerseveranceFrom EverandGrit: The Power of Passion and PerseveranceRating: 4 out of 5 stars4/5 (588)
- Shoe Dog: A Memoir by the Creator of NikeFrom EverandShoe Dog: A Memoir by the Creator of NikeRating: 4.5 out of 5 stars4.5/5 (537)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersFrom EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersRating: 4.5 out of 5 stars4.5/5 (344)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureFrom EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureRating: 4.5 out of 5 stars4.5/5 (474)
- Her Body and Other Parties: StoriesFrom EverandHer Body and Other Parties: StoriesRating: 4 out of 5 stars4/5 (821)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)From EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Rating: 4.5 out of 5 stars4.5/5 (121)
- The Emperor of All Maladies: A Biography of CancerFrom EverandThe Emperor of All Maladies: A Biography of CancerRating: 4.5 out of 5 stars4.5/5 (271)
- The Little Book of Hygge: Danish Secrets to Happy LivingFrom EverandThe Little Book of Hygge: Danish Secrets to Happy LivingRating: 3.5 out of 5 stars3.5/5 (400)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyFrom EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyRating: 3.5 out of 5 stars3.5/5 (2259)
- The Yellow House: A Memoir (2019 National Book Award Winner)From EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Rating: 4 out of 5 stars4/5 (98)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaFrom EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaRating: 4.5 out of 5 stars4.5/5 (266)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryFrom EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryRating: 3.5 out of 5 stars3.5/5 (231)
- Team of Rivals: The Political Genius of Abraham LincolnFrom EverandTeam of Rivals: The Political Genius of Abraham LincolnRating: 4.5 out of 5 stars4.5/5 (234)
- On Fire: The (Burning) Case for a Green New DealFrom EverandOn Fire: The (Burning) Case for a Green New DealRating: 4 out of 5 stars4/5 (73)
- The Unwinding: An Inner History of the New AmericaFrom EverandThe Unwinding: An Inner History of the New AmericaRating: 4 out of 5 stars4/5 (45)
- Fascial Manipulation For Internal Dysfunctions FMIDDocument8 pagesFascial Manipulation For Internal Dysfunctions FMIDdavidzhouNo ratings yet
- Cover Letter 1Document1 pageCover Letter 1api-253411445No ratings yet
- Homework 3Document14 pagesHomework 3api-253411445No ratings yet
- PRFDDocument10 pagesPRFDapi-253411445No ratings yet
- HW 4Document10 pagesHW 4api-253411445No ratings yet
- Homework 1Document14 pagesHomework 1api-253411445No ratings yet
- Homework 2Document19 pagesHomework 2api-253411445No ratings yet
- s516 14s-Homework-4Document4 pagess516 14s-Homework-4api-253411445No ratings yet
- s516 14s-Homework-3Document5 pagess516 14s-Homework-3api-253411445No ratings yet
- Figures and TablesDocument4 pagesFigures and Tablesapi-253411445No ratings yet
- s516 14s-Homework-1Document5 pagess516 14s-Homework-1api-253411445No ratings yet
- Resume 1Document2 pagesResume 1api-253411445No ratings yet
- m1100 s12 v1.1f en - Fender - DesignDocument48 pagesm1100 s12 v1.1f en - Fender - Designdzul fiqarNo ratings yet
- Production of Hard Grade Bitumen For Using in High Modulus Asphalt ConcreteDocument18 pagesProduction of Hard Grade Bitumen For Using in High Modulus Asphalt ConcretePanjiNo ratings yet
- Deflection of BeamsDocument109 pagesDeflection of BeamsNadir Khattak Jr.100% (1)
- MT-LXX Parallel Manual PDFDocument120 pagesMT-LXX Parallel Manual PDFJunior Alexander Rodriguez Rodriguez0% (1)
- L071ME4182DFADocument81 pagesL071ME4182DFAmegamech23No ratings yet
- Translating Mathematical PhrasesDocument16 pagesTranslating Mathematical PhrasesApple Jean Yecyec AlagNo ratings yet
- Grade9 Physics PDFDocument2 pagesGrade9 Physics PDFRajNo ratings yet
- Network Fault Conditions - PPT 3 PDFDocument78 pagesNetwork Fault Conditions - PPT 3 PDFbik_mesiloveNo ratings yet
- Immediate Settlement Analysis Using Finite Element Analysis Models of Fb-MultipierDocument22 pagesImmediate Settlement Analysis Using Finite Element Analysis Models of Fb-MultipierRaaf RifandiNo ratings yet
- G12 NCERT Unit-14 BiomoleculesDocument15 pagesG12 NCERT Unit-14 Biomoleculescherryakila05No ratings yet
- ProjectDocument10 pagesProjectabdul basitNo ratings yet
- VP5 Datasheet Infusion Pump PDFDocument2 pagesVP5 Datasheet Infusion Pump PDFsafouen karrayNo ratings yet
- Body FluidsDocument85 pagesBody FluidsShanta BharNo ratings yet
- Design and Simulation of A QCA 2 To 1 MultiplexerDocument5 pagesDesign and Simulation of A QCA 2 To 1 MultiplexerzubifayazNo ratings yet
- 2021 SEM 4 CC 9 OrganicDocument3 pages2021 SEM 4 CC 9 OrganicGaurav KumarNo ratings yet
- Table and Graph CriteriaDocument3 pagesTable and Graph Criteriaapi-271250844No ratings yet
- Handheld Vital Signs Monitor: XH-30 SeriesDocument4 pagesHandheld Vital Signs Monitor: XH-30 SeriesTopan AssyNo ratings yet
- Friction Clutches PDFDocument14 pagesFriction Clutches PDFamanmittal0850% (2)
- Dewatering Screens: Single-Deck Twin VibratorDocument8 pagesDewatering Screens: Single-Deck Twin Vibratorekrem0867No ratings yet
- Smart Security Camera System For Video Surveillance Using Open CVDocument6 pagesSmart Security Camera System For Video Surveillance Using Open CVlambanaveenNo ratings yet
- Ekg 8Document2 pagesEkg 8Arun SNo ratings yet
- Dw384 Spare Part ListDocument5 pagesDw384 Spare Part ListLuis Manuel Montoya RiveraNo ratings yet
- Class Progress Chart Electrical Installation and Maintenance NC Ii (196 HRS)Document2 pagesClass Progress Chart Electrical Installation and Maintenance NC Ii (196 HRS)Shairrah Claire Bañares BatangueNo ratings yet
- Bsm2300a SeriesDocument324 pagesBsm2300a SeriesIBRAHIMNo ratings yet
- Ratio Worksheet AKDocument12 pagesRatio Worksheet AKChika AuliaNo ratings yet
- Bearing Reliability Improvement-SKFDocument55 pagesBearing Reliability Improvement-SKFAbdulrahman AlkhowaiterNo ratings yet
- Statics and Mechanics of Materials 3Rd Edition Full ChapterDocument41 pagesStatics and Mechanics of Materials 3Rd Edition Full Chapterjason.rabil982100% (29)
- Electricity Markets PDFDocument2 pagesElectricity Markets PDFAhmed KhairiNo ratings yet
- Schiotz TonometerDocument9 pagesSchiotz TonometerDR_LUBYNo ratings yet