Professional Documents
Culture Documents
Gut Microbiota
Uploaded by
Rachel RiosOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Gut Microbiota
Uploaded by
Rachel RiosCopyright:
Available Formats
Int J Clin Exp Med 2016;9(6):10923-10930
www.ijcem.com /ISSN:1940-5901/IJCEM0023128
Original Article
Differential expression of intestinal microbiota in
colorectal cancer compared with healthy controls:
a systematic review and meta-analysis
Haining Liu1, Hao Wu1, Enkhnaran Bilegsaikhan1, Eric Xiaofeng Lu2, Xizhong Shen1,3, Taotao Liu1
1
Department of Gastroenterology, Zhongshan Hospital, Fudan University, Shanghai, China; 2Department of Clini-
cal Medicine, Shanghai Medical College, Fudan University, Shanghai, China; 3Shanghai Institute of Liver Diseases,
Zhongshan Hospital, Fudan University, Shanghai, China
Received January 2, 2016; Accepted March 22, 2016; Epub June 15, 2016; Published June 30, 2016
Abstract: The purpose of this study was to review the differential expression of intestinal microbiota in colorectal
cancer (CRC) with quantitative real-time PCR (q-PCR). An online search within PubMed, Web of Science and Wan-
fang Database up to June 6, 2015 was conducted. We used an original assessment tool to assess the quality of
included articles. A systematic review and meta-analysis described the altered intestinal microbiota in colorectal
cancer versus healthy control (HC). Six articles, involving 192 CRC patients and 264 healthy controls were included.
Scores of quality assessment of these articles ranged from 7 to 16. Significant differences of microbial expression
in CRC compared with HC were found in Bifidobacterium (SMD=-3.303, P=0.048), Faecalibacterium prausnitzii
(SMD=-0.33, P=0.018) and Enterobacteriaceae (SMD=2.69, P=0.009), while total bacteria, Lactobacillus, Bacte-
roides-Prevotella group and Escherichia coli failed to display significant difference. We can conclude that there is
down regulation of some colonization bacteria such as Bifidobacterium and Faecalibacterium prausnitzii, and up
regulation of Enterobacteriaceae in CRC. Microbiota changes also participate in the pathogenesis of CRC.
Keywords: Intestinal microbiota, microbial expression, colorectal cancer, systematic review, meta-analysis
Introduction tional culture-based methods, new molecular
techniques, such as fluorescence in situ hybrid-
Colorectal cancer (CRC), also named large ization (FISH), DNA microarrays, denaturing gra-
bowel cancer, has the third highest cancer mor- dient gel electrophoresis (DGGE), real-time
bidity in the world [1]. The risk factors of quantitative PCR (q-PCR), and pyrosequencing
colorectal cancer include poor eating habits, method, have been applied by scores of studies
genetic susceptibility, colorectal adenoma evo- [5]. Real-time quantitative PCR is a quantitative
lution, and chronic inflammatory stimulation, experiment technique with much higher accu-
etc [2]. In addition, intestinal microbiota are racy relative to previous qualitative or semi-
considered to have an impact on the pathogen- quantitative methods. Compared with pyrose-
esis of CRC as well. Many studies have shown quencing, the new generation of sequencing
direct contact between microbiota and intesti- technique, q-PCR is much cheaper and
nal mucosal cells, wherein some bacteria are becomes a validation tool after pyrosequencing
closely relate to the formation and develop- screening.
ment of CRC, while the metabolites of other
bacteria provide a protective effect to mucosal After reviewing several studies investigating the
cells of the large bowel [3]. abundant variations in bacterial flora, we found
many differences and even contradictions
Intestinal microbiota contain up to 1014 bacte- among the studies. Based on the advantages of
ria [4]. In recent decades, scientists have q-PCR, we retrieved studies using q-PCR and
invented various methods to investigate the performed a systematic review and meta-analy-
microbial community structure. Besides tradi- sis in order to confirm the differential expres-
Altered intestinal microbiota in colorectal cancer
data concerning microbiota;
(2) studies had already been
reported by identical authori-
ties; (3) intervention studies,
animal studies, letters and
reviews.
Quality assessment
We formulated an original as-
sessment tool as there were
no appropriate quality assess-
ment tools for these laborato-
ry studies. We used the follow-
ing twelve criteria based on
diagnostic tests as elaborated
by Bossuyt et al [6]: (1) De-
scribe the study population:
the inclusion and exclusion cri-
teria, setting and locations
where data were collected. (2)
Describe participant sampling:
was the study population a
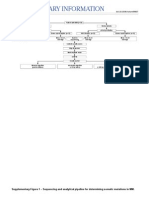
Figure 1. Study selection.
consecutive or random series
of participants? (3) Describe
the diagnostic standard. (4)
sion of intestinal microbiota in colorectal can- Describe the primers and their references. (5)
cer (CRC). Describe technical specifications of methods
involved including how and when measure-
Materials and methods ments were taken, and/or cite references
for index tests and reference standard. (6)
Search strategy Describe methods for calculating test repro-
ducibility, if done. (7) Report when the study
We retrieved articles from PubMed using key- was done, including beginning and ending
words “(“Colorectal Neoplasms” [Mesh] OR dates of recruitment. (8) Report clinical and
“colorectal cancer” OR “colorectal carcinoma”) demographic characteristics of the study popu-
AND (qPCR OR q-PCR OR real-time) AND (micro- lation (e.g., age, sex, comorbidity, recruitment
biome OR microbiota OR flora)”, and retrieved centers). (9) Report the number of participants
articles from Web of Science and Wanfang satisfying the criteria for inclusion that did or
Database using keywords “(“Colorectal Neo- did not undergo the index tests and/or the ref-
plasms” OR “colorectal cancer” OR “colorectal erence standard; describe why participants
carcinoma”) AND (qPCR OR q-PCR OR real- failed to receive either test. (10) Report the
time) AND (microbiome OR microbiota OR distribution of the test results by the results of
flora))” with an end date of June 6, 2015. All the reference standard for continuous results.
references of the studies included were also (11) Report every sample’s expression data or
carefully scrutinized. scatter chart. (12) Report how indeterminate
results, missing responses, and outliers of
Inclusion criteria and exclusion criteria the index tests were handled. Studies were
assigned scores to each criterion by: (1) com-
For a study to be included in this systematic
pletely satisfactory; (2) partially satisfactory; (3)
review, several criteria had to be met: (1) stud-
unsatisfactory or unclear.
ies had to be microbiota studies in CRC versus
healthy controls; (2) samples had to use feces; Data extraction
(3) the methods implemented had to be quanti-
tative real-time PCR techniques. Exclusion cri- Two reviewers independently extracted the fol-
teria were: (1) studies lacked an abundance of lowing data from all eligible studies: (1) basic
10924 Int J Clin Exp Med 2016;9(6):10923-10930
Altered intestinal microbiota in colorectal cancer
Table 1. Characteristics of the 6 studies included in the systematic review
Case/Control Experiment methods
Study Year Region Male ratio Fluorescent q-PCR Reference
Number Mean age
(%) dye instrument
Mira-Pascual L et al 2015 Spain 7/10 71.1/52.6 100/60 SYBR Green I Light Cyder [9]
Miao HF et al 2014 China 19/19 58.0/54.0 47/58 SYBR Green I ABI 7300 [10]
Dong Y et al 2014 China 30/30 49/46 50/50 SYBR Green I ABI 7500 [11]
Wang T et al 2012 China 46/56 60/49a 52/48 SYBR Green I / [12]
Sobhani I et al 2011 France 60/119 67.1/55.8 52/46 / ABI 7000 [13]
Guo SK et al 2010 China 30/30 65.4/60.5 53/57 SYBR Green I Light Cyder [14]
a
Median age.
Table 2. Scores of the 6 studies included using above assessment Wanfang. After removing 14
tool duplicate records, 237 were
Author Year 1 2 3 4 5 6 7 8 9 10 11 12 Score
reserved. Finally, 6 articles
met our inclusion criteria
Mira-Pascual L et al 2015 2 2 2 2 2 2 0 2 0 2 0 0 16
after eliminating animal stud-
Miao HF et al 2014 1 0 0 2 1 0 0 1 0 2 0 0 7 ies and reviews (Figure 1). A
Dong Y et al 2014 1 0 0 1 1 2 2 1 0 2 0 0 10 total of 192 CRC patients
Wang T et al 2012 1 0 2 2 2 2 0 2 0 2 0 0 13 and 264 healthy controls
Sobhani I et al 2011 1 0 0 2 2 0 2 1 0 2 0 1 11 were included.
Guo SK et al 2010 1 0 2 2 1 0 2 1 0 2 0 0 11
No/unclear =0 points; partial =1 point; complete =2 points. Population characteristics
and experiment methods
were listed in Table 1. Four
characteristics of studies, including name of studies were from Asia, while two studies were
the first author, year of publication, country of from Europe. Most of studies used SYBR Green
origin, sample size, mean age, male to female I as fluorescent dye, which had a low specificity
ratio, diagnostic criteria; (2) experimental meth- than TaqMan Probe.
ods: fluorescent dye, q-PCR instrument; (3)
effect size of microbiota: mean (or median), Methodological quality assessment
standard deviation (or quartile) and P value.
We assessed the selected studies with the
Means and standard deviations were estimat-
aforementioned twelve criterion assessment
ed with the method proposed by Stela PH et al
tool. Scores of each study were listed in Table
[7], if the expressions were shown by medians 2. Scores of quality assessment of these arti-
and quartiles. cles ranged from 7 to 16.
Statistical analysis
Microbiota expression
Meta-analysis methods were used to assess 6 kinds of bacteria and total bacteria in 5 arti-
the standardized mean difference (SMD) of cles were included in the meta-analysis. The
bacterial expressions. The random-effect mo- results were expressed as the logarithmic num-
del was applied. The degree of heterogeneity
ber of bacteria per gram stool. Sample sizes,
was quantified by I2 test. Level of significance
means, standard deviations and P values of
was set at P<0.05. The effect of publication
Lactobacillus, Bifidobacterium, Bacteroides-
bias was tested by Egger bias [8]. All analysis
Prevotella group, Faecalibacterium prausnitzii,
used Stata 12.0 software.
Escherichia coli, Enterobacteriaceae and total
Results bacteria are shown in Table 3.
Study selection and characteristics Total bacteria, Escherichia coli and Entero-
bacteriaceae in CRC patients were up-regulat-
251 records were identified from the databas- ed, while only Enterobacteriaceae (SMD=2.69,
es, of which 18 were from PubMed, 31 were P=0.009, 95% CI=0.66 to 4.72) had the signifi-
from Web of Science and 204 were from cantly changed expression (Figure 2). Lactoba-
10925 Int J Clin Exp Med 2016;9(6):10923-10930
Altered intestinal microbiota in colorectal cancer
Table 3. Effect size and P value of various bacteria
CRC Healthy Control
Bacteria Author Year P value
N Mean Std N Mean Std
Total bacteria Mira-Pascual L et al 2015 7 11.2 0.20 9 10.88 0.20 /
Miao HF et al 2014 19 11.4 0.80 19 11.5 0.50 0.954
Sobhani I et al 2011 60 11.8 0.56 119 11.88 0.35 0.21
Lactobacillus Mira-Pascual L et al 2015 7 6.59 1.16 9 5.93 0.24 /
Dong Y et al 2014 30 6.54 0.43 30 9.53 0.74 0.076
Sobhani I et al 2011 60 9.56 0.85 119 9.56 0.95 0.27
Guo SK et al 2010 30 4.52 0.49 30 9.25 0.83 0.00
Bifidobacterium Mira-Pascual L et al 2015 7 8.6 0.34 9 9.09 0.32 /
Dong Y et al 2014 30 6.54 0.43 30 9.53 0.74 0.082
Sobhani I et al 2011 60 9.89 1.06 119 9.85 1.22 0.9
Guo SK et al 2010 30 4.52 0.49 30 9.25 0.83 0.02
Bacteroides-Prevotella group Mira-Pascual L et al 2015 7 10.36 0.34 9 10.4 0.26 /
Dong Y et al 2014 30 9.83 0.53 30 11.14 0.57 0.009
Sobhani I et al 2011 60 10.76 0.55 119 10.48 0.83 0.009
Faecalibacterium prausnitzii Mira-Pascual L et al 2015 7 8.19 0.24 9 8.15 0.25 /
Miao HF et al 2014 19 8.9 1.70 19 9.6 0.90 0.35
Sobhani I et al 2011 60 10.75 1.02 119 11.04 0.80 0.72
Escherichia coli Sobhani I et al 2011 60 8.14 1.34 119 8.14 1.28 0.25
Guo SK et al 2010 30 5.82 0.47 30 4.68 0.32 0.01
Enterobacteriaceae Mira-Pascual L et al 2015 7 8.25 0.48 9 7.41 0.55 /
Dong Y et al 2014 30 10.59 0.63 30 8.51 0.49 0.047
Enterococcus faecalis Guo SK et al 2010 30 10.6 0.3 30 4.95 0.24 0.00
Enterococcus faecium Guo SK et al 2010 30 5.74 0.16 30 5.03 0.43 0.00
Fusobacterium Miao HF et al 2014 19 9.1 1.40 19 7.2 1.50 0.002
Eubacterium rectale Miao HF et al 2014 19 9.2 1.20 19 9.2 1.30 0.840
cillus, Bifidobacterium, Bacteroides-Prevotella play an essential role in the mechanisms of
group and Faecalibacterium prausnitzii in CRC obesity, diabetes mellitus, irritable bowel syn-
patients were down-regulated, while Bifido- drome (IBS), inflammatory bowel disease (IBD)
bacterium (SMD=-3.303, P=0.048, 95% CI=- and colorectal cancer [15]. Some evidence sug-
6.57 to -0.03) and Faecalibacterium prausnitzii gested that the differential expression of bacte-
(SMD=-0.33, P=0.018, 95% CI=-0.60 to -0.05) ria and tumorigenesis of the large bowel is a
had the significantly different expression relationship of reciprocal causation. A certain
(Figure 3). combination of life styles, microbiota and their
metabolites leads to intestinal inflammation.
In addition, Enterococcus faecalis, Enteroco- The study by Junhai Ou et al linked different
ccus faecium, Fusobacterium and Eubacterium CRC incidence and microbiota to distinct
rectale were reported to be differently ex- dietary habits of rural Africans and African
pressed in CRC patients from that in the healthy Americans [16]. On the other hand, CRC per-
controls in one single article. tained as a systemic disease releases cyto-
kines into circulation and influences the micro-
Publication bias
environment of intestines, leading to changes
Egger bias test did not show evident publica- in the microbial community structure.
tion bias with the P value of 0.179 (Figure 4).
In this study, we observed a significant
Discussion decreased expression in Bifidobacterium and
Faecalibacterium prausnitzii, as well as a sig-
Recently, abundant scientific and clinical stud- nificant increased expression in Enterobac-
ies have identified that intestinal microbiota teriaceae. According to the results, we specu-
10926 Int J Clin Exp Med 2016;9(6):10923-10930
Altered intestinal microbiota in colorectal cancer
Figure 2. Forest plot of up-regulated bacteria including total bacteria, Escherichia coli and Enterobacteriaceae.
Figure 3. Forest plot of down-regulated bacteria including Lactobacillus, Bifidobacterium, Bacteroides-Prevotella
group and Faecalibacterium prausnitzii.
10927 Int J Clin Exp Med 2016;9(6):10923-10930
Altered intestinal microbiota in colorectal cancer
quantification methods of
DNA standard samples were
different in included studies.
DNA quantification using ul-
traviolet spectrophotometer
in early studies [14] contrib-
uted the largest errors in this
study.
We tried to discuss the con-
sequences of altered micro-
biota from the perspective of
mechanisms. A decrease in
Bifidobacterium, one of the
most crucial colonization
bacteria, leads to immuno-
deficiency of hosts, which
allows tumor cells to escape
Figure 4. Egger’s funnel plot of various bacteria. from immune surveillance
[17]. Faecalibacterium prau-
snitzii, a member of Clostri-
late that Lactobacillus failed to reveal a signifi- dium cluster IV, is one of the main butyrate pro-
cant difference due to inadequate sample size; ducers. Butyrate can be absorbed and utilized
however, total bacteria, Bacteroides-Prevotella by intestinal mucosal cells, and then inhibit
and Escherichia Coli did not display a funda- pro-inflammatory mediators such as tumor
mental association with CRC. Otherwise, nu- necrosis factor (TNF) and IL-6 [3]. In addition,
merous bacteria such as Enterococcus faeca- Faecalibacterium prausnitzii is an anti-inflam-
lis, Enterococcus faecium and Fusobacterium matory bacterium with secreted metabolites
were observed to have significant increases, which can block nuclear factor κB activation
but were not included in the meta-analysis due and IL-8 production [18]. Enterobacteriaceae,
to the limited number of studies available. In including Enterobacter, Escherichia, Klebsiella,
short, it can be determined that there is down- Salmonella, Shigella and Yersinia, are patho-
regulation of normal bacterial flora and up-reg- genic or conditional pathogenic species which
ulation of pathogenic bacteria in those with would reproduce at greater rates when coloni-
CRC. zation bacterial flora is decreased. Some gen-
era of Enterobacteriaceae can produce poly-
The high heterogeneity of meta-analysis cannot amines to enhance their invasiveness. Oxidative
be ignored; therefore we should be conscien- stress as the result of high levels of polyamines
tious in accepting the synthetic result. A sub- is associated with CRC [19, 20]. Enterococcus
group analysis, sensitivity analysis or meta- faecalis can induce oxidative DNA damage
and mitochondrial dysfunction through reactive
regression was not appropriate because the
oxygen species (ROS) in epithelial cells [21].
number of included studies was less than 10.
Fusobacterium nucleatum can stimulate the
For our part, the heterogeneity was derived
proliferation of CD11b+ myeloid cells and over-
from experimental methodology. Although
expression of pro-inflammatory factors like
experimenters used the same q-PCR tech-
Ptgs2, Scyb1, Tnf and Mmp. These pro-inflam-
nique, primers, and Fluorescent dye, the stor-
matory factors belong to NF-κB signaling path-
age of fecal samples and the quantity of DNA way, which leads to the tumorigenesis of epi-
standard samples might account for errors. The thelial cells [22, 23].
storage condition of fecal samples prior to stor-
age at -80°C was uncontrollable. Under pro- In summary, we had identified the association
longed exposure at room temperature, the between the differential expression of intesti-
reproduction of aerobic bacteria may have nal microbiota and colorectal cancer through a
caused damage to DNA of anaerobic bacteria, meta-analysis of 5 case-control studies, and
which are the main flora in the large bowel. The had discussed its corresponding mechanisms.
10928 Int J Clin Exp Med 2016;9(6):10923-10930
Altered intestinal microbiota in colorectal cancer
Further studies with larger samples and more ses of controlled trials with binary endpoints.
species/clusters of bacteria or fungi are need- Stat Med 2006; 25: 3443-3457.
ed to identify the association and mechanisms [9] Mira-Pascual L, Cabrera-Rubio R, Ocon S, Cos-
are needed to be elaborated. tales P, Parra A, Suarez A, Moris F, Rodrigo L,
Mira A and Collado MC. Microbial mucosal co-
Acknowledgements lonic shifts associated with the development
of colorectal cancer reveal the presence of dif-
The authors would like to thank the members of ferent bacterial and archaeal biomarkers. J
Prof. Xi-Zhong Shen’s Laboratory for their help- Gastroenterol 2015; 50: 167-179.
ful discussion and critical reading of the manu- [10] Miao H, Wu N, Luan C, Yang X, Zhang R, Lv N
script. This study was supported by National and Zhu B. Quantitation of intestinal Fusobac-
Nature Science Foundation of China (No. terium and butyrate-producing bacteria in pa-
81000968; No. 81101540; No. 81101637; tients with colorectal adenomas and colorectal
cancer. Acta Microbiologica Sinica 2014; 54:
No. 81172273; No. 81272388; No. 81301820,
1228-1234.
No. 81472673), Doctoral Fund of Ministry of [11] Dong Y, He X, Yu Z, Li J and Zhang A. Character-
Education of China (20120071110058), The istics of patients with colorectal cancer using
National Clinical Key Special Subject of China. real-time PCR and PCR-DGGE. Chinese Journal
of Microbiology and Immunology 2014: 551-
Disclosure of conflict of interest 553.
[12] Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X,
None.
Jia W, Cai S and Zhao L. Structural segregation
Address correspondence to: Dr. Taotao Liu, De- of gut microbiota between colorectal cancer
partment of Gastroenterology, Zhongshan Hospital, patients and healthy volunteers. ISME J 2012;
6: 320-329.
Fudan University, Room 207, Building 3, No. 180
[13] Sobhani I, Tap J, Roudot-Thoraval F, Roperch
Fenglin Road, Shanghai 200032, China. Tel:
JP, Letulle S, Langella P, Corthier G, Tran VN
+86-21-64041990-2070; Fax: +86-21-64432583; and Furet JP. Microbial dysbiosis in colorectal
E-mail: liu.taotao@zs-hospital.sh.cn cancer (CRC) patients. PLoS One 2011; 6:
e16393.
References [14] Guo S, Bao W, Gong K, Shao J, Chen D and
Wang K. SYBR green I real-time fluorescence
[1] Jemal A, Bray F, Center MM, Ferlay J, Ward E quantitative pcr analysis of variation of intesti-
and Forman D. Global cancer statistics. CA nal microflora in patients with colorectal can-
Cancer J Clin 2011; 61: 69-90.
cer. Chinese Journal of Bases and Clinics in
[2] Townsend CM, Beauchamp RD, Evers BM and
General Surgery 2010; 17: 463-468.
Mattox KL. Sabiston textbook of surgery. 19.
[15] Aziz Q, Dore J, Emmanuel A, Guarner F and
Amsterdam: Elsevier; 2012. pp. 1337-1345.
Quigley EM. Gut microbiota and gastrointesti-
[3] Louis P, Hold GL and Flint HJ. The gut microbi-
nal health: current concepts and future direc-
ota, bacterial metabolites and colorectal can-
tions. Neurogastroenterol Motil 2013; 25:
cer. Nat Rev Microbiol 2014; 12: 661-672.
[4] Savage DC. Microbial ecology of the gastroin- 4-15.
testinal tract. Annu Rev Microbiol 1977; 31: [16] Ou J, Carbonero F, Zoetendal EG, DeLany JP,
107-133. Wang M, Newton K, Gaskins HR and O’Keefe
[5] Taverniti V and Guglielmetti S. Methodological SJ. Diet, microbiota, and microbial metabolites
issues in the study of intestinal microbiota in in colon cancer risk in rural Africans and Afri-
irritable bowel syndrome. World J Gastroenter- can Americans. Am J Clin Nutr 2013; 98: 111-
ol 2014; 20: 8821-8836. 120.
[6] Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis [17] Iwashita J, Sato Y, Sugaya H, Takahashi N, Sa-
CA, Glasziou PP, Irwig LM, Moher D, Rennie D, saki H and Abe T. mRNA of MUC2 is stimulated
de Vet HC and Lijmer JG. The STARD statement by IL-4, IL-13 or TNF-alpha through a mitogen-
for reporting studies of diagnostic accuracy: activated protein kinase pathway in human
explanation and elaboration. Clin Chem 2003; colon cancer cells. Immunol Cell Biol 2003;
49: 7-18. 81: 275-282.
[7] Hozo SP, Djulbegovic B and Hozo I. Estimating [18] Sokol H, Pigneur B, Watterlot L, Lakhdari O,
the mean and variance from the median, Bermudez-Humaran LG, Gratadoux JJ, Blugeon
range, and the size of a sample. BMC Med Res S, Bridonneau C, Furet JP, Corthier G, Grang-
Methodol 2005; 5: 13. ette C, Vasquez N, Pochart P, Trugnan G, Thom-
[8] Harbord RM, Egger M and Sterne JA. A modi- as G, Blottiere HM, Dore J, Marteau P, Seksik P
fied test for small-study effects in meta-analy- and Langella P. Faecalibacterium prausnitzii is
10929 Int J Clin Exp Med 2016;9(6):10923-10930
Altered intestinal microbiota in colorectal cancer
an anti-inflammatory commensal bacterium [22] Kostic AD, Chun E, Robertson L, Glickman JN,
identified by gut microbiota analysis of Crohn Gallini CA, Michaud M, Clancy TE, Chung DC,
disease patients. Proc Natl Acad Sci U S A Lochhead P, Hold GL, El-Omar EM, Brenner D,
2008; 105: 16731-16736. Fuchs CS, Meyerson M and Garrett WS. Fuso-
[19] Di Martino ML, Campilongo R, Casalino M, Mi- bacterium nucleatum potentiates intestinal
cheli G, Colonna B and Prosseda G. Poly- tumorigenesis and modulates the tumor-im-
amines: emerging players in bacteria-host in- mune microenvironment. Cell Host Microbe
teractions. Int J Med Microbiol 2013; 303: 2013; 14: 207-215.
484-491. [23] Kostic AD, Gevers D, Pedamallu CS, Michaud
[20] Pegg AE. Toxicity of polyamines and their meta- M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ,
bolic products. Chem Res Toxicol 2013; 26: Tabernero J, Baselga J, Liu C, Shivdasani RA,
1782-1800. Ogino S, Birren BW, Huttenhower C, Garrett WS
[21] Strickertsson JA, Rasmussen LJ and Friis-Han- and Meyerson M. Genomic analysis identifies
sen L. Enterococcus faecalis infection and re- association of Fusobacterium with colorectal
active oxygen species down-regulates the miR- carcinoma. Genome Res 2012; 22: 292-298.
17-92 cluster in gastric adenocarcinoma cell
culture. Genes (Basel) 2014; 5: 726-738.
10930 Int J Clin Exp Med 2016;9(6):10923-10930
You might also like
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeFrom EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeRating: 4 out of 5 stars4/5 (5794)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreFrom EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreRating: 4 out of 5 stars4/5 (1090)
- Never Split the Difference: Negotiating As If Your Life Depended On ItFrom EverandNever Split the Difference: Negotiating As If Your Life Depended On ItRating: 4.5 out of 5 stars4.5/5 (838)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceFrom EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceRating: 4 out of 5 stars4/5 (894)
- Grit: The Power of Passion and PerseveranceFrom EverandGrit: The Power of Passion and PerseveranceRating: 4 out of 5 stars4/5 (587)
- Shoe Dog: A Memoir by the Creator of NikeFrom EverandShoe Dog: A Memoir by the Creator of NikeRating: 4.5 out of 5 stars4.5/5 (537)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureFrom EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureRating: 4.5 out of 5 stars4.5/5 (474)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersFrom EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersRating: 4.5 out of 5 stars4.5/5 (344)
- Her Body and Other Parties: StoriesFrom EverandHer Body and Other Parties: StoriesRating: 4 out of 5 stars4/5 (821)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)From EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Rating: 4.5 out of 5 stars4.5/5 (119)
- The Emperor of All Maladies: A Biography of CancerFrom EverandThe Emperor of All Maladies: A Biography of CancerRating: 4.5 out of 5 stars4.5/5 (271)
- The Little Book of Hygge: Danish Secrets to Happy LivingFrom EverandThe Little Book of Hygge: Danish Secrets to Happy LivingRating: 3.5 out of 5 stars3.5/5 (399)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyFrom EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyRating: 3.5 out of 5 stars3.5/5 (2219)
- The Yellow House: A Memoir (2019 National Book Award Winner)From EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Rating: 4 out of 5 stars4/5 (98)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaFrom EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaRating: 4.5 out of 5 stars4.5/5 (265)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryFrom EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryRating: 3.5 out of 5 stars3.5/5 (231)
- Team of Rivals: The Political Genius of Abraham LincolnFrom EverandTeam of Rivals: The Political Genius of Abraham LincolnRating: 4.5 out of 5 stars4.5/5 (234)
- On Fire: The (Burning) Case for a Green New DealFrom EverandOn Fire: The (Burning) Case for a Green New DealRating: 4 out of 5 stars4/5 (73)
- The Unwinding: An Inner History of the New AmericaFrom EverandThe Unwinding: An Inner History of the New AmericaRating: 4 out of 5 stars4/5 (45)
- 【Zybio】IFU-Lepgen 96Document59 pages【Zybio】IFU-Lepgen 96Daniel Huachani CoripunaNo ratings yet
- Richter 2017Document10 pagesRichter 2017Rachel RiosNo ratings yet
- Renal Congestion in HFDocument9 pagesRenal Congestion in HFRachel RiosNo ratings yet
- Biomolecules 05 02223Document24 pagesBiomolecules 05 02223Rachel RiosNo ratings yet
- Thrombosis Risk Assessment As A Guide To Quality Patient CareDocument9 pagesThrombosis Risk Assessment As A Guide To Quality Patient CareRachel RiosNo ratings yet
- Acid Base Balance in DMDocument9 pagesAcid Base Balance in DMRachel RiosNo ratings yet
- Handbook: For Clinical Management of DengueDocument124 pagesHandbook: For Clinical Management of DengueraattaiNo ratings yet
- Immuno TherapyDocument120 pagesImmuno TherapyRachel RiosNo ratings yet
- H.pylory InCAPDDocument4 pagesH.pylory InCAPDRachel RiosNo ratings yet
- Kuliah Perlemakan Hati 2012Document17 pagesKuliah Perlemakan Hati 2012Rachel RiosNo ratings yet
- Allo HSCTDocument8 pagesAllo HSCTRachel RiosNo ratings yet
- BPCO - FabbriDocument41 pagesBPCO - FabbriRachel RiosNo ratings yet
- (Involvement of The Skin-Mucosal Tissues) : 1.mildDocument4 pages(Involvement of The Skin-Mucosal Tissues) : 1.mildRachel RiosNo ratings yet
- AcsDocument11 pagesAcsRachel RiosNo ratings yet
- 1 s2.0 S0422763813003026 Main PDFDocument13 pages1 s2.0 S0422763813003026 Main PDFRachel RiosNo ratings yet
- 08e00833 PDFDocument89 pages08e00833 PDFRachel RiosNo ratings yet
- 1 s2.0 S0422763813003026 Main PDFDocument13 pages1 s2.0 S0422763813003026 Main PDFRachel RiosNo ratings yet
- 210 FullDocument9 pages210 FullRachel RiosNo ratings yet
- Adenoca EsofagusDocument7 pagesAdenoca EsofagusRachel RiosNo ratings yet
- EsophagealDocument3 pagesEsophagealRachel RiosNo ratings yet
- Tumor EsophagealDocument4 pagesTumor EsophagealRachel RiosNo ratings yet
- Adenokarsinoma EsofagusDocument8 pagesAdenokarsinoma EsofagusRachel RiosNo ratings yet
- Adenocarcinoma EsophagealDocument12 pagesAdenocarcinoma EsophagealRachel RiosNo ratings yet
- Adenokarsinoma EsofagusDocument5 pagesAdenokarsinoma EsofagusRachel RiosNo ratings yet
- What is the difference between UNG and UDG in qPCRDocument3 pagesWhat is the difference between UNG and UDG in qPCRbdd.budNo ratings yet
- Food Safety and Quality ReferEnce Guide - BIORADDocument52 pagesFood Safety and Quality ReferEnce Guide - BIORADAhsanal KasasiahNo ratings yet
- 23.-Eperytrozoonosis en CerdosDocument10 pages23.-Eperytrozoonosis en Cerdoscyntia medina de martinezNo ratings yet
- Liu2017 EhpDocument6 pagesLiu2017 EhpmokengNo ratings yet
- Alfaro-Fernández, Ana - 2016Document7 pagesAlfaro-Fernández, Ana - 2016NoemiNo ratings yet
- Microbiology Uncertainty of Measurement According To ISO 19036-MatDocument51 pagesMicrobiology Uncertainty of Measurement According To ISO 19036-MatoktaNo ratings yet
- Postharvest Biology and Technology: Ya Zhong Jin, de Qing LV, Wen Wei Liu, Hong Yan Qi, Xiao Hang BaiDocument9 pagesPostharvest Biology and Technology: Ya Zhong Jin, de Qing LV, Wen Wei Liu, Hong Yan Qi, Xiao Hang BaiBiSii OanaaNo ratings yet
- (Pds Patklin) : Perhimpunan Dokter Spesialis Patologi Klinik Dan Kedokteran Laboratorium IndonesiaDocument13 pages(Pds Patklin) : Perhimpunan Dokter Spesialis Patologi Klinik Dan Kedokteran Laboratorium IndonesiaNida ChoerunnisaNo ratings yet
- S3018E Manual-Mycobacterium Tuberculosis DNA Fluorescence Diagnostic Kit (PCR-Fluorescence Probing) V03-20190929Document3 pagesS3018E Manual-Mycobacterium Tuberculosis DNA Fluorescence Diagnostic Kit (PCR-Fluorescence Probing) V03-20190929paulaNo ratings yet
- Olerup QTYPE HLA Typing Kits IFU v9-3Document19 pagesOlerup QTYPE HLA Typing Kits IFU v9-3huripNo ratings yet
- A New Tool For CRISPR-Cas13a-Based Cancer Gene TherapyDocument14 pagesA New Tool For CRISPR-Cas13a-Based Cancer Gene TherapySharon TribhuvanNo ratings yet
- Handbook of New Technologies For Genetic Improvement of Legumes (CRC 2008)Document688 pagesHandbook of New Technologies For Genetic Improvement of Legumes (CRC 2008)Senjuti Sinharoy100% (1)
- TFS-Assets LSG Manuals MAN0006735 AmpliSeq DNA RNA LibPrep UGDocument90 pagesTFS-Assets LSG Manuals MAN0006735 AmpliSeq DNA RNA LibPrep UGcoutinhobvictorNo ratings yet
- Initial Genome Sequencing and Analysis of Multiple Myeloma - SuppDocument75 pagesInitial Genome Sequencing and Analysis of Multiple Myeloma - SuppShantanu KumarNo ratings yet
- A Mass Sacrifice of Children and CamelidDocument15 pagesA Mass Sacrifice of Children and CamelidMaría Del Carmen Espinoza CórdovaNo ratings yet
- Resume - Tran Dinh Quang LocDocument1 pageResume - Tran Dinh Quang LocTony TranNo ratings yet
- Speedy Assay Product List May 2017Document4 pagesSpeedy Assay Product List May 2017bhbdNo ratings yet
- (MiCo BioMed) Molecular Diagnostics For COVID-19 - Eng - 0324 - DecDocument40 pages(MiCo BioMed) Molecular Diagnostics For COVID-19 - Eng - 0324 - Decdiogenes amaroNo ratings yet
- Id Pipeta Ep-5Document39 pagesId Pipeta Ep-5erikaNo ratings yet
- General Katalog PT. AmoebaDocument23 pagesGeneral Katalog PT. AmoebaMulyanaNo ratings yet
- 308-1127 - Nucleic Acid-Based Techniques-AmplificationDocument11 pages308-1127 - Nucleic Acid-Based Techniques-AmplificationmeiNo ratings yet
- Micropropagación de Glossonema VariansDocument16 pagesMicropropagación de Glossonema VariansmatateteNo ratings yet
- Rates of Dark CO2 Fixation Are Driven by Microbial Biomass in A Temperate Forest SoilDocument58 pagesRates of Dark CO2 Fixation Are Driven by Microbial Biomass in A Temperate Forest SoilIsais NNo ratings yet
- AriaMx DataSheet 5991-5711ENDocument2 pagesAriaMx DataSheet 5991-5711ENbotond77No ratings yet
- Alkhidmat Diagnostic Center Blood Bank: Virology & Genetic LabDocument1 pageAlkhidmat Diagnostic Center Blood Bank: Virology & Genetic LabNum RahNo ratings yet
- Ecprm 09 00577Document21 pagesEcprm 09 00577HETA JOSHINo ratings yet
- Spek Dan Gambar Lelang 2Document3 pagesSpek Dan Gambar Lelang 2Fajar Pandu WijayaNo ratings yet
- NewEarth - University COVID-19 Intelligence BriefDocument47 pagesNewEarth - University COVID-19 Intelligence BriefTony Lambert100% (1)
- zhang-et-al-2023-the-newest-oxford-nanopore-r10-4-1-full-length-16s-rrna-sequencing-enables-the-accurate-resolution-ofDocument20 pageszhang-et-al-2023-the-newest-oxford-nanopore-r10-4-1-full-length-16s-rrna-sequencing-enables-the-accurate-resolution-ofScientific PeopleNo ratings yet