Professional Documents
Culture Documents
Yeast Genetic Analyses
Uploaded by
Patrycja PietruszkaOriginal Description:
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Yeast Genetic Analyses
Uploaded by
Patrycja PietruszkaCopyright:
Available Formats
7 Genetic Analyses
7.1 Overviews with Examples
There are numerous approaches for the isolation and characterization of mutations in yeast.
Generally, a haploid strain is treated with a mutagen, such as ethylmethanesulfonate, and the
desired mutants are detected by any one of a number of procedures. For example, if Yfg-
(Your Favorite Gene) represents an auxotrophic requirement, such as arginine, or
temperature-sensitive mutants unable to grow at 37°C, the mutants could be scored by replica
plating. Once identified, the Yfg- mutants could be analyzed by a variety of genetic and
molecular methods. Three major methods, complementation, meiotic analysis and molecular
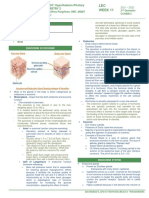
cloning are illustrated in Figure 7.1.
Genetic complementation is carried out by crossing the Yfg- MATa mutant to each of the
tester strains MATα yfg1, MATα yfg2, etc., as well as the normal control strain MATα .
These yfg1, yfg2, etc., are previously defined mutations causing the same phenotype. The
diploid crosses are isolated and the Yfg trait is scored. The Yfg+ phenotype in the
heterozygous control cross establishes that the Yfg- mutation is recessive. The Yfg- phenotype
in MATα yfg1 cross, and the Yfg+ phenotype in the MATα yfg2, MATα yfg3, etc., crosses
reveals that the original Yfg- mutant contains a yfg1 mutation.
Meiotic analysis can be used to determine if a mutation is an alteration at a single genetic
locus and to determine genetic linkage of the mutation both to its centromere and to other
markers in the cross. As illustrated in Figure 7.1, the MATa yfg1 mutant is crossed to a normal
MATα strain. The diploid is isolated and sporulated. Typically, sporulated cultures contain
the desired asci with four spores, as well as unsporulated diploid cells and rare asci with less
than four spores. The sporulated culture is treated with snail extract which contains an enzyme
that dissolves the ascus sac, but leaves the four spores of each tetrad adhering to each other. A
portion of the treated sporulated culture is gently transferred to the surface of a petri plate or
an agar slab. The four spores of each cluster are separated with a microneedle controlled by a
micromanipulator. After separation of the desired number of tetrads, the ascospores are
allowed to germinate and form colonies on complete medium. The haploid segregants can
then be scored for the Yfg+ and Yfg- phenotypes. Because the four spores from each tetrad are
the product of a single meiotic event, a 2:2 segregation of the Yfg+:Yfg- phenotypes is
indicative of a single gene. If other markers are present in the cross, genetic linkage of the
yfg1 mutation to the other markers or to the centromere of its chromosome could be revealed
from the segregation patterns.
The molecular characterization of the yfg1 mutation can be carried out by cloning the wild-
type YFG1+ gene by complementation, as illustrated in Figure 7.1 and described below
(Section 11.1 Cloning by Complementation).
You might also like
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryFrom EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryRating: 3.5 out of 5 stars3.5/5 (231)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)From EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Rating: 4.5 out of 5 stars4.5/5 (119)
- Never Split the Difference: Negotiating As If Your Life Depended On ItFrom EverandNever Split the Difference: Negotiating As If Your Life Depended On ItRating: 4.5 out of 5 stars4.5/5 (838)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaFrom EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaRating: 4.5 out of 5 stars4.5/5 (265)
- The Little Book of Hygge: Danish Secrets to Happy LivingFrom EverandThe Little Book of Hygge: Danish Secrets to Happy LivingRating: 3.5 out of 5 stars3.5/5 (399)
- Grit: The Power of Passion and PerseveranceFrom EverandGrit: The Power of Passion and PerseveranceRating: 4 out of 5 stars4/5 (587)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyFrom EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyRating: 3.5 out of 5 stars3.5/5 (2219)
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeFrom EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeRating: 4 out of 5 stars4/5 (5794)
- Morgellons Disease Related To Black Magic Rituals PDFDocument30 pagesMorgellons Disease Related To Black Magic Rituals PDFadu66680% (5)
- Team of Rivals: The Political Genius of Abraham LincolnFrom EverandTeam of Rivals: The Political Genius of Abraham LincolnRating: 4.5 out of 5 stars4.5/5 (234)
- Shoe Dog: A Memoir by the Creator of NikeFrom EverandShoe Dog: A Memoir by the Creator of NikeRating: 4.5 out of 5 stars4.5/5 (537)
- The Emperor of All Maladies: A Biography of CancerFrom EverandThe Emperor of All Maladies: A Biography of CancerRating: 4.5 out of 5 stars4.5/5 (271)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreFrom EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreRating: 4 out of 5 stars4/5 (1090)
- Her Body and Other Parties: StoriesFrom EverandHer Body and Other Parties: StoriesRating: 4 out of 5 stars4/5 (821)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersFrom EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersRating: 4.5 out of 5 stars4.5/5 (344)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceFrom EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceRating: 4 out of 5 stars4/5 (890)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureFrom EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureRating: 4.5 out of 5 stars4.5/5 (474)
- The Unwinding: An Inner History of the New AmericaFrom EverandThe Unwinding: An Inner History of the New AmericaRating: 4 out of 5 stars4/5 (45)
- The Yellow House: A Memoir (2019 National Book Award Winner)From EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Rating: 4 out of 5 stars4/5 (98)
- On Fire: The (Burning) Case for a Green New DealFrom EverandOn Fire: The (Burning) Case for a Green New DealRating: 4 out of 5 stars4/5 (73)
- Clinical Chemistry 2 LectureDocument46 pagesClinical Chemistry 2 LectureGillian Joy Pagaling100% (1)
- Wechsler Intelligence Scale For ChildrenDocument4 pagesWechsler Intelligence Scale For ChildrenCyndi Whitmore67% (3)
- Anxiety DisordersDocument20 pagesAnxiety DisordersAngela BautistaNo ratings yet
- ABO, RH, Minor Blood Grps AHG Test Pretransfusion Test Automation in BBDocument32 pagesABO, RH, Minor Blood Grps AHG Test Pretransfusion Test Automation in BBJill Arciaga0% (1)
- Thyroid UltrasoundDocument62 pagesThyroid UltrasoundYoungFanjiens100% (1)
- Pet ScanDocument40 pagesPet ScanAkhilesh KhobragadeNo ratings yet
- NephronDocument5 pagesNephronrdeguzma100% (1)
- Innate and Adaptive Immunity: - Innate Is FIRST LINE OF DEFENCE: No Prior Exposure Needed - Comprised OfDocument31 pagesInnate and Adaptive Immunity: - Innate Is FIRST LINE OF DEFENCE: No Prior Exposure Needed - Comprised OfBegumHazinNo ratings yet
- Lecture Planner - Zoology - Yakeen NEET 3.0 2024Document6 pagesLecture Planner - Zoology - Yakeen NEET 3.0 2024RyzoxbeatsNo ratings yet
- M.03 Non-Mendelian GeneticsDocument5 pagesM.03 Non-Mendelian Geneticsno veNo ratings yet
- Corey Dambacher, Ph.D. Recognized As A Professional of The Year by Strathmore's Who's Who Worldwide PublicationDocument2 pagesCorey Dambacher, Ph.D. Recognized As A Professional of The Year by Strathmore's Who's Who Worldwide PublicationPR.comNo ratings yet
- Mercury Poisoning Symptoms and TreatmentsDocument1 pageMercury Poisoning Symptoms and TreatmentsRakheeb BashaNo ratings yet
- Module For Students 1 - 2 - 3-1 EditDocument15 pagesModule For Students 1 - 2 - 3-1 EditJanuardi IndraNo ratings yet
- eenMedInfo Store PDFDocument183 pageseenMedInfo Store PDFJeanne De Wet50% (2)
- Simina Maria Boca, PH.D.: EducationDocument13 pagesSimina Maria Boca, PH.D.: EducationRohit AroraNo ratings yet
- Clinical and Etiological Profile of Unprovoked Thrombosis in Young Patients Admitted at A Tertiary Care HospitalDocument5 pagesClinical and Etiological Profile of Unprovoked Thrombosis in Young Patients Admitted at A Tertiary Care HospitalfarhanomeNo ratings yet
- Mother Touch in Periodontal Therapy: Chorion MembraneDocument7 pagesMother Touch in Periodontal Therapy: Chorion MembraneInternational Journal of Innovative Science and Research TechnologyNo ratings yet
- What is Gene TherapyDocument2 pagesWhat is Gene TherapyNarasimha MurthyNo ratings yet
- Parkinsonism (巴金森氏症) and other movement disordersDocument29 pagesParkinsonism (巴金森氏症) and other movement disordersShing Ming TangNo ratings yet
- Definition and Nature of ResearchDocument20 pagesDefinition and Nature of ResearchJiggs LimNo ratings yet
- L96 - Patna Lab 2 (Home Visit) : Patientreportscsuperpanel - SP - General - Template01 - SC (Version: 7)Document3 pagesL96 - Patna Lab 2 (Home Visit) : Patientreportscsuperpanel - SP - General - Template01 - SC (Version: 7)Rohit Gautam DwivediNo ratings yet
- Comprehensive Guide to E@J StandardsDocument46 pagesComprehensive Guide to E@J StandardsEdson LimaNo ratings yet
- Comparative Evaluation of Alternate Emergency FixativesDocument14 pagesComparative Evaluation of Alternate Emergency FixativesIJAR JOURNALNo ratings yet
- Lecture On Physiology of HaemoglobinDocument26 pagesLecture On Physiology of HaemoglobinMubasharAbrarNo ratings yet
- Methemoglobinemia Diagnosis & Treatment GuideDocument11 pagesMethemoglobinemia Diagnosis & Treatment GuideAranaya DevNo ratings yet
- European Vol-32 No-6 2011Document15 pagesEuropean Vol-32 No-6 2011mahaberani_zNo ratings yet
- AutismDocument30 pagesAutismLIWANGA LIWANGANo ratings yet
- Biology Half Yearly 12Document14 pagesBiology Half Yearly 12Tarun MonNo ratings yet
- CRL 9609Document3 pagesCRL 9609naveenmi2No ratings yet