Professional Documents
Culture Documents
Tail PCR Protocol
Uploaded by
Felipe ZapataOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Tail PCR Protocol
Uploaded by
Felipe ZapataCopyright:
Available Formats
TAIL (Thermal Asymmetric Interlaced) PCR 1
Andrew Clarke
Allan WIlson Centre for |olecular Ecology and EvolutIon
|assey UnIversIty
NEW ZEALAN0
Created: 19/06/200J
Last updated: 10/10/200J
TAIL (THERMAL ASYMMETRIC INTERLACED) PCR
for obtaining DNA sequence from flanking regions
Adapted from Liu, Y.-G. & Whittier, R. F. (1995)
Thermal Asymmetric Interlaced PCR: Automatable amplification and sequencing of
insert end fragments from P1 and YAC clones for chromosome walking. Genomics
25: 674-681.
1. Primer Design
Design three adjacent primers from your sequence (priming outwards from the
sequence). These become the specific (SP) primers. SP1 is the innermost primer,
SP2 is the middle primer and SP3 is nearest the end of the known sequence, and are
used in the 1, 2 and 3 PCR reactions respectively.
The SP primers need to be ~20-mers and have a T
m
of 60-62C.
Use the following arbitrary degenerate (AD) primers.
Name Sequence (5-3) Length T
m
Degeneracy
TAIL-AD1 NGTCGASWGANAWGAA 16-mer 46C 128
TAIL-AD2 GTNCGASWCANAWGTT 16-mer 46C 128
TAIL-AD3 WGTGNAGWANCANAGA 16-mer 45C 256
S = C or G
W = A or T
Choose one AD primer (e.g. AD1) and use it for the 1, 2 and 3 reactions. If this
primer fails try AD2, and so on. More bands can be expected when using AD3
(because of the higher degeneracy).
SP1 SP2 SP3
= known sequence
= unknown sequence
= specific primer
TAIL (Thermal Asymmetric Interlaced) PCR 2
2. Primary (1) PCR Reaction
Set up the 1 PCR reaction (20L volume).
1
Milli-Q H
2
O 0.3
Betaine (5M) 4.0 (1M)
dNTPs (2mM) 2.5 (0.25mM)
10 PCR Buffer 2.0 (1)
MgCl
2
(25mM) 1.0 (1.25mM)
AD Primer* (10pmol/L) 8.0 (4M)
SP1 Primer (10pmol/L) 1.0 (0.5M)
Genomic DNA 1.0
Taq Polymerase (5U/L) 0.2 (1U)
TOTAL 20 L
* Use AD primer AD1, AD2 or AD3.
Use the following 1 PCR program:
Step
1 94C 2 min 1
2 94C 30 sec
3 60C 1 min 5
4 72C 2 min
5 94C 30 sec
6 25C 1 sec 1
7* 72C 2 min
8 94C 30 sec
9 44C 1 min 10
10 72C 2 min
11 94C 30 sec
12 60C 1 min
13 72C 2 min
14 94C 30 sec
15 60C 1 min 15 supercycles
16 72C 2 min
17 94C 30 sec
18 44C 1 min
19 72C 2 min
20 72C 5 min 1
21 4C HOLD
* Ramp from 25C to 72C over 3 min (47C = +0.25C/s). All other ramping
speeds are limited to 1C/s.
TAIL (Thermal Asymmetric Interlaced) PCR 3
3. Secondary (2) PCR Reaction
Dilute the 1 PCR reaction 100 with MilliQ-H
2
O. Set up the 2 PCR reaction (20L
volume).
1
Milli-Q H
2
O 2.3
Betaine (5M) 4.0 (1M)
dNTPs (2mM) 2.5 (0.25mM)
10 PCR Buffer 2.0 (1)
MgCl
2
(25mM) 1.0 (1.25mM)
AD Primer* (10pmol/L) 6.0 (3M)
SP2 Primer (10pmol/L) 1.0 (0.5M)
1 PCR Product (diluted 100) 1.0
Taq Polymerase (5U/L) 0.2 (1U)
TOTAL 20 L
* Use the same AD primer as in the 1 reaction.
Use the following 2 PCR program:
Step
1 94C 2 min 1
2 94C 30 sec
3 60C 1 min
4 72C 2 min
5 94C 30 sec
6 60C 1 min 15 supercycles
7 72C 2 min
8 94C 30 sec
9 44C 1 min
10 72C 2 min
11 72C 5 min 1
12 4C HOLD
Ramping speed should be limited to 1C/s.
TAIL (Thermal Asymmetric Interlaced) PCR 4
4. Tertiary (3) PCR Reaction
Dilute the 2 PCR reaction 100 with MilliQ-H
2
O. Set up the 3 PCR reaction (20L
volume).
1
Milli-Q H
2
O 4.3
Betaine (5M) 4.0 (1M)
dNTPs (2mM) 2.5 (0.25mM)
10 PCR Buffer 2.0 (1)
MgCl
2
(25mM) 1.0 (1.25mM)
AD Primer* (10pmol/L) 4.0 (2M)
SP3 Primer (10pmol/L) 1.0 (0.5M)
2 PCR Product (diluted 100) 1.0
Taq Polymerase (5U/L) 0.2 (1U)
TOTAL 20 L
* Use the same AD primer as in the 1 and 2 reaction.
Use the same PCR program as for the 2 PCR reaction.
5. Agarose Electrophoresis of Products
Run the 1, 2 and 3 products in adjacent lanes on a 1.5% agarose gel (5L each of
the 1 and 2 reactions and all 20L of the 3 reaction). Look for the characteristic
shift in size of the desired product. The change in product size should correspond to
the length between SP primers.
If the 3 reaction contains just one product a PCR cleanup can be performed and the
product sequenced directly (using the SP3 primer and/or the AD primer). More often
there are multiple 3 products, in which case the desired band can be excised from
agarose and gel extracted, before being sequenced directly.
If direct sequencing is problematic the product can be cloned and sequenced.
You might also like
- High TAIL PCRDocument5 pagesHigh TAIL PCRChristianFaltadoCantosNo ratings yet
- The Polymerase Chain ReactionDocument35 pagesThe Polymerase Chain ReactionAthika Darumas PutriNo ratings yet
- Bisulfite Conversion and MSP ProtocolDocument2 pagesBisulfite Conversion and MSP Protocolitaimo100% (1)
- TEV Protease Purification ProtocolDocument7 pagesTEV Protease Purification ProtocolChen GuttmanNo ratings yet
- BST DNA/RNA PolymeraseDocument2 pagesBST DNA/RNA PolymeraseSBS GenetechNo ratings yet
- Intro To NGS - Torsten Seemann - PeterMac - 27 Jul 2012Document51 pagesIntro To NGS - Torsten Seemann - PeterMac - 27 Jul 2012Gusti Deky Genetika ScienceNo ratings yet
- 2-14 Epoxidation of AlkenesDocument2 pages2-14 Epoxidation of AlkenesMohd Zulhelmi AzmiNo ratings yet
- SOAL P13-5 (Elements of Chemical Reaction Engineering 4 Edition Foggler)Document31 pagesSOAL P13-5 (Elements of Chemical Reaction Engineering 4 Edition Foggler)Adilla PratiwiNo ratings yet
- GREDocument7 pagesGREBoris ShalomovNo ratings yet
- Determination of ParacetamolDocument4 pagesDetermination of ParacetamolChandan SinghNo ratings yet
- Isolation of Curcumin From TurmericDocument2 pagesIsolation of Curcumin From TurmericyadranNo ratings yet
- Introduction MTT AssayDocument10 pagesIntroduction MTT Assay16_dev5038No ratings yet
- Solutions For Reactor KineticsDocument2 pagesSolutions For Reactor Kineticszy_yfNo ratings yet
- SP LAB Experiment ManualDocument69 pagesSP LAB Experiment ManualChemGuyNo ratings yet
- Cells Culture Counting, Dilution and Plating CalculatorDocument1 pageCells Culture Counting, Dilution and Plating CalculatoritaimoNo ratings yet
- Plasmid DNA Quantification of DNADocument3 pagesPlasmid DNA Quantification of DNAHuishin LeeNo ratings yet
- Lab Report Cstr-Intro Appa ProceDocument6 pagesLab Report Cstr-Intro Appa Procesolehah misniNo ratings yet
- High-Throughput Droplet Digital PCR System For Absolute PDFDocument7 pagesHigh-Throughput Droplet Digital PCR System For Absolute PDFLuong Nguyen100% (1)
- AOAC Official Method 980.13 Fructose, Glucose, Lactose, Maltose, and Sucrose in Milk ChocolateDocument2 pagesAOAC Official Method 980.13 Fructose, Glucose, Lactose, Maltose, and Sucrose in Milk ChocolateBrian MontoyaNo ratings yet
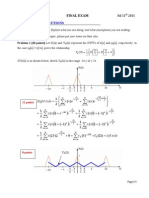
- Final Exam 2012Document12 pagesFinal Exam 2012Mat MorashNo ratings yet
- Dna Sequencing by Sanger MethodDocument19 pagesDna Sequencing by Sanger MethodSethu SKNo ratings yet
- Improved HPLC Method For The Determination of Curcumin, Demethoxycurcumin, and BisdemethoxycurcuminDocument5 pagesImproved HPLC Method For The Determination of Curcumin, Demethoxycurcumin, and BisdemethoxycurcuminWidiyastuti DarwisNo ratings yet
- Chomczynski, Mackey - 1995 - Short Technical Reports. Modification of The TRI Reagent Procedure For Isolation of RNA From PolysaccharideDocument6 pagesChomczynski, Mackey - 1995 - Short Technical Reports. Modification of The TRI Reagent Procedure For Isolation of RNA From PolysaccharideskljoleNo ratings yet
- (EE332) (09ECE) (Group13) Report ProjectDocument23 pages(EE332) (09ECE) (Group13) Report ProjectNgô ĐạtNo ratings yet
- GCMS-QP2010 User'sGuide (Ver2.5) PDFDocument402 pagesGCMS-QP2010 User'sGuide (Ver2.5) PDFnguyenvietanhbtNo ratings yet
- RNA EditingDocument19 pagesRNA Editingrag.1607No ratings yet
- Application Report DMA 80 Evo Coal USREV061019Document4 pagesApplication Report DMA 80 Evo Coal USREV061019Roni GustiwaNo ratings yet
- Hair Dye Chemical SensitizationDocument18 pagesHair Dye Chemical SensitizationSarah CarrollNo ratings yet
- A TpoDocument4 pagesA TpoJimboreanu György PaulaNo ratings yet
- Dopamine Drug Monograph YOR-PD-025Document2 pagesDopamine Drug Monograph YOR-PD-025Andy AdibNo ratings yet
- Protein Extraction BufferDocument1 pageProtein Extraction BufferRabia AnjumNo ratings yet
- Active Site TitrationDocument4 pagesActive Site TitrationokpitNo ratings yet
- Knauer - Food PreservativesDocument1 pageKnauer - Food PreservativesMaryanta HadisudiroNo ratings yet
- Digital PCR ProtocolDocument11 pagesDigital PCR ProtocolitaimoNo ratings yet
- MMPVXXX Installation Manual (En HW) PDFDocument35 pagesMMPVXXX Installation Manual (En HW) PDFeves5No ratings yet
- Merck Colunas HPLCDocument94 pagesMerck Colunas HPLCMileane BuschNo ratings yet
- Polymerase Chain ReactionDocument14 pagesPolymerase Chain ReactionDavidMugambi100% (1)
- Sop of Alkaline PhosphataseDocument6 pagesSop of Alkaline PhosphataseUMMID WashimNo ratings yet
- Digital Signal Processing LabDocument26 pagesDigital Signal Processing LabDurotan123No ratings yet
- W2 SolutionsDocument3 pagesW2 Solutionsjohn smither100% (1)
- Team 3Document21 pagesTeam 3lux0008No ratings yet
- UAH - Fall 2014 - ISE 690 - Helvaci - Final Exam Formula SheetDocument2 pagesUAH - Fall 2014 - ISE 690 - Helvaci - Final Exam Formula SheetAnaan EemusNo ratings yet
- Homework 7 Bme260 s16 KeyDocument4 pagesHomework 7 Bme260 s16 Keyxryceu100% (2)
- Safety Assignment 1Document5 pagesSafety Assignment 1Nurwani HussinNo ratings yet
- Solved Examples For Chapter 19Document7 pagesSolved Examples For Chapter 19SofiaNo ratings yet
- Synthesis, Characterization and Pharmacological Evaluation of Some Cinnoline DerivativesDocument6 pagesSynthesis, Characterization and Pharmacological Evaluation of Some Cinnoline DerivativesIOSRjournalNo ratings yet
- EE341 FinalDocument4 pagesEE341 FinalLe HuyNo ratings yet
- PCR PresentationDocument24 pagesPCR PresentationsandrapaolamtzfNo ratings yet
- PCR and Agarose Gel ElectrophoresisDocument5 pagesPCR and Agarose Gel ElectrophoresisEamon Barkhordarian100% (1)
- Material Balance of Styrene Production PDocument12 pagesMaterial Balance of Styrene Production PSteve WanNo ratings yet
- SDS-PAGE Tris-Tricine SystemDocument2 pagesSDS-PAGE Tris-Tricine SystemChuong GioNo ratings yet
- Solved - A Carnot Refrigerator Has Tetrafluoroethane As The Work...Document4 pagesSolved - A Carnot Refrigerator Has Tetrafluoroethane As The Work...MingNo ratings yet
- User ManualDocument26 pagesUser ManualRodney CutinhoNo ratings yet
- Vasicek Model SlidesDocument10 pagesVasicek Model SlidesnikhilkrsinhaNo ratings yet
- MKBS221. PRAC 5 REPORT Last OneDocument11 pagesMKBS221. PRAC 5 REPORT Last OneNOMKHULEKO ALICENo ratings yet
- PCR CalculationsDocument7 pagesPCR Calculationsmarwatsum100% (1)
- M MLV RTDocument4 pagesM MLV RTZNo ratings yet
- PCR Labwork 2 ENGDocument4 pagesPCR Labwork 2 ENGmigas1996No ratings yet
- Gene Specific RTPCRDocument4 pagesGene Specific RTPCRQuimico Inmunologia GeneticaNo ratings yet
- Thermo Scientific Taq Dna Polymerase With KCL Buffer: DescriptionDocument3 pagesThermo Scientific Taq Dna Polymerase With KCL Buffer: DescriptionCHIRANJEEVINo ratings yet
- Retropharyngeal Abscess (RPA)Document15 pagesRetropharyngeal Abscess (RPA)Amri AshshiddieqNo ratings yet
- Muet Topic 10 City Life Suggested Answer and IdiomsDocument3 pagesMuet Topic 10 City Life Suggested Answer and IdiomsMUHAMAD FAHMI BIN SHAMSUDDIN MoeNo ratings yet
- Achievement Test Science 4 Regular ClassDocument9 pagesAchievement Test Science 4 Regular ClassJassim MagallanesNo ratings yet
- ICT ContactCenterServices 9 Q1 LAS3 FINALDocument10 pagesICT ContactCenterServices 9 Q1 LAS3 FINALRomnia Grace DivinagraciaNo ratings yet
- EarthmattersDocument7 pagesEarthmattersfeafvaevsNo ratings yet
- Lecture 8: Separation DesignDocument45 pagesLecture 8: Separation DesignRavi Kiran MNo ratings yet
- Hedayati2014 Article BirdStrikeAnalysisOnATypicalHeDocument12 pagesHedayati2014 Article BirdStrikeAnalysisOnATypicalHeSharan KharthikNo ratings yet
- Evolution of Fluidized Bed TechnologyDocument17 pagesEvolution of Fluidized Bed Technologyika yuliyani murtiharjonoNo ratings yet
- PCB Engraver Operator Manual PDFDocument41 pagesPCB Engraver Operator Manual PDFmyoshkeuNo ratings yet
- Labor Case DigestDocument2 pagesLabor Case DigestJhollinaNo ratings yet
- Monitor Zoncare - PM-8000 ServicemanualDocument83 pagesMonitor Zoncare - PM-8000 Servicemanualwilmer100% (1)
- Annual Sustainability Report 2022-23 FinalDocument93 pagesAnnual Sustainability Report 2022-23 FinalLakshay JajuNo ratings yet
- Mapagbigay PT1 Group1Document4 pagesMapagbigay PT1 Group1Hazel SarmientoNo ratings yet
- Medicina 57 00032 (01 14)Document14 pagesMedicina 57 00032 (01 14)fauzan nandana yoshNo ratings yet
- Listes de Produits GAURAPADDocument1 pageListes de Produits GAURAPADBertrand KouamNo ratings yet
- Peseshet - The First Female Physician - (International Journal of Gynecology & Obstetrics, Vol. 32, Issue 3) (1990)Document1 pagePeseshet - The First Female Physician - (International Journal of Gynecology & Obstetrics, Vol. 32, Issue 3) (1990)Kelly DIOGONo ratings yet
- ProjectxDocument8 pagesProjectxAvinash KumarNo ratings yet
- 45relay Rm4ua PDFDocument1 page45relay Rm4ua PDFtamky SubstationNo ratings yet
- Gastroschisis and Omphalocele PDFDocument8 pagesGastroschisis and Omphalocele PDFUtama puteraNo ratings yet
- Sample UploadDocument14 pagesSample Uploadparsley_ly100% (6)
- File3 PDFDocument119 pagesFile3 PDFikkemijnnaam0% (1)
- Gpat Reference NotesDocument9 pagesGpat Reference NotesPreethi KiranNo ratings yet
- Lesson 1 Animal CareDocument8 pagesLesson 1 Animal CareLexi PetersonNo ratings yet
- A0002 HR Operations Manual Quick Reference FileDocument6 pagesA0002 HR Operations Manual Quick Reference FileRaffy Pax Galang RafaelNo ratings yet
- Education - Khóa học IELTS 0đ Unit 3 - IELTS FighterDocument19 pagesEducation - Khóa học IELTS 0đ Unit 3 - IELTS FighterAnna TaoNo ratings yet
- The Nursing ShortageDocument6 pagesThe Nursing Shortageapi-455495817No ratings yet
- Reading Test - 3 Clinical Depression Text ADocument17 pagesReading Test - 3 Clinical Depression Text AJisha JanardhanNo ratings yet
- El MeligyDocument7 pagesEl Meligysystematic reviewNo ratings yet
- Sheehan SyndromeDocument6 pagesSheehan SyndromeArvie TagnongNo ratings yet
- Business-Plan (John Lloyd A Perido Grade 12-Arc)Document4 pagesBusiness-Plan (John Lloyd A Perido Grade 12-Arc)Jaypher PeridoNo ratings yet