Professional Documents
Culture Documents
Swiss Deep View PDF
Uploaded by
Karthi ShanmugamOriginal Description:
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Swiss Deep View PDF
Uploaded by
Karthi ShanmugamCopyright:
Available Formats
INTRODUCTION TO SWISS DEEP VIEW: Understanding the Mechanism of a Serine Protease
References: 1. Lindert, L. M. Protein structure exploration using Swiss PDB Viewer. http://chemweb.calpoly.edu/chem/lindert/PSE/Proteinstructure-FS.htm. (11/15/04). 2. Lindert, L. M. Presented at the 228th National Meeting of the American Chemical Society, Philadelphia, PA, August 2004; paper CHED 11. In previous biochemistry courses you have learned to manipulate threedimensional structures in the web-embedded molecular visualization software Protein Explorer. The purpose of this bioinformatics exercise is to introduce an alternative molecular visualization tool, Swiss Deep View, that offers alternative benefits/features to Deep View. Deep view is a free software tool like Protein Explorer, but it is not webembedded and works well on Mac or PC platforms. PE does not. Finally, we will use Deep View at various points during the term and your efficiency with the use of the program is expected. The present activity has been adapted from Dr. Lisa Lindert and will look at the protein bovine beta trypsin. Once you have learned how to use Swiss Deep View to view structures, you will be able to download all kinds of molecular structures for visualization/manipulation! The free software that will be used for this exercise is available for PC or Mac at: http://www.expasy.org/spdbv/text/download.htm. You may find it helpful throughout the term to have this software loaded on your personal computer to allow you to look at the structures of various biomolecules covered in the course. Instructions for this project are shown below and are based on those found at: . We will be following the basic instructions with modifications. See instructions below.
*if you were not aware how to reference a website, the format shown here is the American Chemical Societys accepted format:
Authors last name, first name, if available. Title of article/page as listed on the site. URL of page. (date accessed)
Analysis of a Serine Protease with Swiss Deep View: Step 1: Find the Protein Data Bank entry 1. Go directly to the Protein Data Bank to find our model structure for this exercise: http://www.rcsb.org/pdb/ 2. In Search the Archive, type in trypsin. How many hits do you get? What kinds of information regarding each of the structures is immediately available? What additional information do you find when you hit the Explore button for any of the listed structures? 3. Find the PDB entry for bovine beta-trypsin, a more narrow search. Now, how many hits do you get? How could you narrow the search to bovine beta-trypsin with an inhibitor? How many hits does that give? 4. Use the structure with PDB ID = 1TPP. The full entry for this protein will be given. What types of information are given (e.g. who were the authors, what was the original citation, how long has this data been posted in PDB, what was the source of the protein, etc)? Give a description of this entry. 5. Click the Display Molecule in the left side panel. The coordinates will download to your computer. This may take a moment. Step 2: Load 1TPP into Deep View 1. Open the Deep View software (aka Swiss PDB-Viewer). Your screen will adjust to 250 colors. 2. In the file menu click on Open and choose the 1TPP file you just downloaded (at this point you could open any particular structural file you had previously downloaded and saved, i.e. other biomolecules we might view in this course!). 3. The structure of 1TPP should appear as a stick model. In addition, a text file will appear on top of the program bar. Close this text window so that what you see is the program bar and the protein structure in the window below the bar. Step 3: Learning to use Deep View Adapted from Lindert (1): Next we will work with this structure. Our initial goal will be the generation of a ribbon diagram and a space-filled diagram that highlights some key structural features of trypsin. It is helpful to be familiar with the basics of the mechanism of serine proteases. Serine proteases, specifically chymotrypsin, is covered in VVP in Chapter 11, pg. 307. These enzymes were covered in great detail in Biochemistry. As a reminder, proteases hydrolyze amide bonds, i.e. they degrade proteins by cutting up the backbone at the amide bond. The mechanism of serine proteases involved a catalytic triad of amino acid residues: Serine (serves as a nucleophile), Histidine (serves as an acid/base) and Aspartic Acid (serves to stabilize charge on His when it is protonated on the imidazole ring). While the mechanisms of all serine proteases are very similar, their specificity differs, that is, some cut-up proteins from the C-terminal end, some cut from the middle of a protein, some prefer to cut near bulky side chains, etc Trypsin cleaves following
basic amino acid residues throughout the protein. In this exercise we will look at the catalytic triad and also examine the structure of a bound protein inhibitor in the active site and understand why it is bound where it is bound and what aspects of the natural substrate it mimics. NOTE: Depending on the speed of your computer and its graphics capabilities you may or may not want to toggle on the 3D rendering options in your window. To turn these features on go to the Display menu and turn on 'Render in Solid 3D' and 'Use OpenGL rendering.' You can play with these options to see which (or both on better computers) works best with your system. If neither works well, turning these features on only when you are finished working with the model (rotating, coloring etc.) or want to briefly see your model will work. Deep View operates in as a series of windows. This is useful because you can open and close the windows as you need them. The main toolbar will always be open. The other windows are opened from the Window menu located at the top right. These will open as independent windows and can be opened and closed at any time.
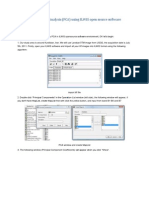
The Control Panel: 1. Open the Control Panel (in the main toolbar, click on Window and then Control Panel. 2. Main Features of the Control Panel are indicated and defined below:
Label Explanation A This check-box toggles the visibility of the structure in this layer B If checked, you can move the structure around C The 'group' column lists the amino acid (in order from n to C terminus) with the heteroatoms at the bottom of the list. To the left of the group colum can also be a column with letters from A-Z. If there is more than one amino acid chain in a molecule, this is shown by these letters. D "show" is short for show backbone. If a "V" is listed (a check in Deep View) in this column then the backbone for that particular atom will be shown E "side" is short for side chain. If this is checked then the side chain for that particular atom will be shown. F "labl" is short for Label. If this is checked the amino acid will be labeled in the structure with the three letter abbreviation and the residue number. These labels can be changed in the Preferences menu G This symbol stands for 'dots surface' or the van der Waals or accessible surface area of the protein. When checked the vdW or accessible surface will be shown. The drop down arrow allows you to select which surface will be displayed. H "ribn" stands for Ribbon. If checked a ribbon will be traced through the backbone and it will be drawn as a helix or strand depending on the classification of that amino acid (see K) I 'col' stands for color. Highlighting particular residues and clicking on this box brings up a dialogue box that allows you to change the color of the selected residue. J Drop down menu for the color option. You use this drop down menu to choose whether you are changing the color of the backbone, backbone+sidechain, sidechain or the ribbon. K The small letters to the left of the group name indicate whether this residue is located in a helix or a strand. These are different from the capital letters that can be located to the left of these letters that indicate a separate chain. Table taken from (1). 4
Keystroke shortcuts for the Control Panel: clicking within a row This checks/unchecks ('V') next to a residue and activates the feature in that column control+click on a group This selects all groups that you click on while control is pressed. This is useful for selecting a group of residues and especially useful if they are not continuous within the sequence. shift+click within a column This checks/unchecks all residues in that column within a layer control+shift+click within a column This checks/unchecks all residues in that column within all layers (if working with multiple layers) Control+Z Undo Table taken from (1).
The Main Toolbar, Rotation, Zooming and Centering The main toolbar contains all the major tools for navigating the structure, moving the structure and changing your perspective. Navigation, Movement and Rotation
Label A B
Description Clicking on this button centers the molecule on the screen Clicking on this button changes your cursor to the 'hand' which allows you to move the entire molecule or selection C Clicking on this button changes your cursor to the zoom-in zoom-out cursor. Dragging your mouse forward and back will zoom you into and out of the structure. D Clicking on this button changes your cursor to the rotate cursor. Moving your mouse side-to-side and up and down will rotate the molecule or selection E Clicking on this changes the rotation from a global rotation or a rotation about the axis of symmetry of the molecule. Often this is useful to play with when you are trying to get a particular angle and can't quite get the structure to move the way you would like. F Toggles between moving the entire structure or just the selected residues. Table taken from (1).
Changing Colors You can change the color of individual residues, ribbons, loops and strucures by selecting them individually and in groups and then changing the color of the ribbon, sidechain, backbone etc. from the Control Panel. There are other options, however that give you important information and are easier to work with than selecting groups within the control panel. These selections are found in the Color drop down menu in the Main Toolbar. The table below describes the most useful features of this menu. In the Color Menu, use the act on selection at the top to toggle what is changed.
Some descriptions/definitions: Label Information A Colors the individual atoms according to a standard color scheme B Colors by the type of amino acid. Non-polar = grey, polar =yellow, acidic = red, basic =blue. C Colors by secondary structural element. Helices are red, sheets yellow and loops white, by default. You may change these colors in the Preferences menu. D Colors by secondary structure succession from N to C terminus. This colors by wavelength of the visible spectrum from N terminus = violet to C terminus =red. Table taken from (1).
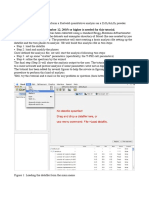
Step 4: Learning more about Deep View Adapted from Lindert (1): The goal of this portion of the tutorial is to get you used to some more advanced features in Deep View. This will allow you to select specific residues and allow you to display only those residues, or to color these differently or display them in a particular representation. You can also compute H-bonds to see which amino acids are connected non-covalently. If you combine these selections, image manipulation and computational tools with the skills you learned above you will be able to create images that convey a lot of structural information. In addition to providing a tool for viewing structures, Deep View also allows you to examine the phi/psi angles of the residues in the protein. In the Window menu is an option to generate a Ramachandran plot for the protein you are viewing. This is a useful tool to find residues that may be strained out of normal, allowed conformations, or to assess the quality of the protein structure (if 90% of the residues have phi/psi angles outside the allowed regions, you may have some doubts!). Slab View This option, located in the Display menu allows you to slice the protein and see successive cross sections through the protein structure. When this option is checked, hold down the shift button on your keyboard and move your mouse back and forth. This will 'slice' the protein into sections that you scroll through with your mouse. In the preferences menu you can change the width of the 'slice' that you take to allow finer and coarser views into the structure. Selecting Specific Residues There are several ways to select residues. In fact, there is a whole menu dedicated to the task of selecting particular residues. This menu offers a variety of ways to select residues. You can then decide to color these a particular color, zoom in to see these, etc. None -- removes all selections. This is useful to start fresh after making some selections. All -- Selects all residues Inverse selection -- select some residues, then choose this to choose all residues but those currently selected. Visible groups -- zoom in and then use this to select those residues you have in the current view. Pick on screen -- choose then and then click on particular residue(s). These will be selected in the control panel. Group Kind -- allows you to select a particular amino acid, nucleic acid, heteroatom, solvent molecule or disulfide bond. GroupProperty -- allows you to select acidic, basic, polar and non-polar amino acids. Secondary Structure -- allows you to select all residues in a beta sheet, alph helix or coils. It also has options to select all non-trans amino acids, amino acids with phi/psi angles outside of core regions or amino acids with phi/psi angles outside of allowed regions.
Several of these options are also located in the main toolbar. The icon with an eye and clock looking circle, located in the main toolbar, allows you to select amino acids within a defined radius, i.e. you could find all the amino acids within 10 of the active site, for instance. You click the icon, click on a residue on the screen and the 'Display Radius' dialog box appears. Try it! The icon with an eye and arrows all pointing to a central dot doesn't do much in the way of selecting in the Control Panel, but if you click this button and then choose an atom in the structure the view will change such that the chosen atom is centered in your view. Try it!
If you would like to identify the residues located next to the inhibitor you have two options.... Option 1. Go to the Select menu. Click on 'Pick on Screen.' Then go on the screen and click on the residues you wish to select. They will be highlighted in red on the control panel. Option 2. Select the inhibitor heteroatom from the Control Panel. Go to the Select menu. Click on 'Neighbors of Selected aa..' then select the radius you wish to observe and the residues within that radius will be selected. If you wish to only see these residues you may deselect all atoms in the structure (remove all checkmarks) and then choose the option to display only those atoms selected. This allows you to see only what is in the neighborhood of your inhibitor in the active site. Combine this with your 'Slab' view abilities and you are dissecting an enzyme active site layer by layer!!!
Now, with the experience using Deep View and loading a protein structure, we will begin the real exercise analyzing the structure of a serine protease! Note: be sure to master these skills as you will see this again both on the mid-term exam and in the lab portion of the course. Project to complete: 1. Create a ribbon diagram of trypsin. Color it by secondary structure and then by secondary structure succession. Remember to turn off the side chains. 2. Place labels at the N-terminal and C-terminal ends. 3. Make the heteroatom appear in ball and stick representation. Use checkmarks in the show and side columns. 4. Go to Select menu and choose select all. Now, rotate the structure so that the bound inhibitor in the active site is visible. You may need to zoom in and out on the structure to get the best view, but keep the whole protein in the frame. 5. Save a copy of this view (see note below on printing). Prepare a printout of this view with a figure caption stating what is shown. 6. Show the structure in space-fill mode, i.e. with van der Waals surface. 7. Color the residues by type. Remember to color both the back bone and side chain. 8. Keep the inhibitor in ball and stick form so it can be seen in the enzyme active site. Make a note of the possible interactions between the inhibitor and the protein that can be seen in this view. 9. Save a copy of this view (see note below on printing). Prepare a printout of this view with a figure caption stating what is shown. 10. Turn on slab mode by going into Display and clicking slab. Hold down Shift and move your mouse forward and back! Cross-sections of the protein now appear. Find a cross-section that clearly shows major interactions between the inhibitor and the residues within the active site. 11. Prepare a slab view with a figure caption stating what is shown. 12. Prepare a Ramachandran plot. What is a Ramachandran plot and what does it indicate about the 1TPP protein structure?
A note on printing from Swiss Deep View with the HHMI laptops: Once you have manipulated the structure to the desired view, press the "prtscrn" function key. The whole display is copied into the clipboard at that point. Now, paste the image into "Paint" (which is under Accessories). Crop and paste the image in Paint to the desired region. Once you have a final version, paste from Paint into Microsoft Word with no distortions of the image. You may then add your captions or other embedded text easily with the image in Word and save as a Word file.
The lab report for this project should include answers to all the questions posed in bold and underlined in the instructions for the project. In addition, the following printouts should be included: Printouts to attach to lab report for Deep View of a Serine Protease: Ribbon diagram with figure caption (one page). Space-filling model with inhibitor visible and with caption (one page). A slab view with inhibitor visible and captions (one page). Ramachandran plot with caption (one page).
10
You might also like
- Protein Structure ExplorationDocument22 pagesProtein Structure Explorationsuveer698No ratings yet
- GFP TutorialDocument19 pagesGFP TutorialLivsNo ratings yet
- PyMol PracticalDocument7 pagesPyMol PracticalTom FlemingNo ratings yet
- Insulina TutorialDocument20 pagesInsulina TutorialLivsNo ratings yet
- PyMOL ProtocolDocument5 pagesPyMOL ProtocolMonica Fan HsuNo ratings yet
- Molecular Biology Basics Tutorial: Design Primers, Add AnnotationsDocument21 pagesMolecular Biology Basics Tutorial: Design Primers, Add AnnotationsDaniela GuardiaNo ratings yet
- Guidelines and QuestionsDocument6 pagesGuidelines and Questionsd nNo ratings yet
- Discovery Studio 2.1 Tutorials GuideDocument130 pagesDiscovery Studio 2.1 Tutorials Guidedark_geneNo ratings yet
- Introduction to Discovery Studio VisualizerDocument31 pagesIntroduction to Discovery Studio VisualizerbitMorph3rNo ratings yet
- Argus Molecular Docking 1Document5 pagesArgus Molecular Docking 1escherichioNo ratings yet
- Project MEGA ProtocolDocument5 pagesProject MEGA ProtocolSaneet AgrawalNo ratings yet
- Biochemistry ResearchDocument14 pagesBiochemistry ResearchArya ChowdhuryNo ratings yet
- Rast TutorialDocument10 pagesRast TutorialPeter JustinNo ratings yet
- Introduction to Bioinformatics DatabasesDocument10 pagesIntroduction to Bioinformatics DatabasesRaj Kumar SoniNo ratings yet
- Moe TutorDocument66 pagesMoe TutorPrasanna BabuNo ratings yet
- Assignment 1ADocument12 pagesAssignment 1AianNo ratings yet
- Tutorial RNA-Seq Analysis Part 1Document8 pagesTutorial RNA-Seq Analysis Part 1Lee LI PINNo ratings yet
- Pelajaran 4Document6 pagesPelajaran 4nazkasihNo ratings yet
- Skyline Targeted Method Editing TutorialDocument26 pagesSkyline Targeted Method Editing TutorialCristian BorosNo ratings yet
- Manual XJDocument26 pagesManual XJmiguelNo ratings yet
- Aspen TutorialDocument33 pagesAspen TutorialSarah RasheedNo ratings yet
- Discovery Studio VisualizerDocument31 pagesDiscovery Studio VisualizerShin KaojuNo ratings yet
- DipTrace TutorialDocument134 pagesDipTrace TutorialMetalloyNo ratings yet
- Bioinformatics Workshop LDH Worksheet-1Document4 pagesBioinformatics Workshop LDH Worksheet-1alem010No ratings yet
- BLAST - Practic InformationDocument2 pagesBLAST - Practic InformationSarah JuárezNo ratings yet
- Protein structure modification using SPDBVDocument10 pagesProtein structure modification using SPDBVPrayukta PadelkarNo ratings yet
- Lab 3 - Multiple Sequence Alignment: Bioinformatic Methods I Lab 3Document14 pagesLab 3 - Multiple Sequence Alignment: Bioinformatic Methods I Lab 3Ariadna Andrade AlvaradoNo ratings yet
- Dock and Score Tyrosine Kinase InhibitorsDocument14 pagesDock and Score Tyrosine Kinase InhibitorsvarinderkumarscribdNo ratings yet
- (Directions Modified From Sarah Deel's Bio 125 Directions) : Week 7: Sequence AnalysisDocument7 pages(Directions Modified From Sarah Deel's Bio 125 Directions) : Week 7: Sequence Analysisjj1064No ratings yet
- PIPE2 TutorialDocument11 pagesPIPE2 TutorialWolelaw EndalewNo ratings yet
- PyMOL Tutorial JKLDocument18 pagesPyMOL Tutorial JKLShubham James BasuNo ratings yet
- PyMOL TutorialDocument7 pagesPyMOL TutorialMatheus SilvaNo ratings yet
- Tutorial PymolDocument16 pagesTutorial PymolfahraniNo ratings yet
- Skyline Hi-Res MetabolomicsDocument18 pagesSkyline Hi-Res MetabolomicsShahinuzzamanAdaNo ratings yet
- Classification and Clustering of Animals Using Decision Trees and K-MeansDocument4 pagesClassification and Clustering of Animals Using Decision Trees and K-MeansKaustubh KhareNo ratings yet
- KnowYourMolecule ChEMBL KNIMEDocument11 pagesKnowYourMolecule ChEMBL KNIMEMarcos FerreiraNo ratings yet
- Flowjo-Guide - EMBLDocument11 pagesFlowjo-Guide - EMBLMasse Ak-FaNo ratings yet
- SOP UV-VIS Spectrophotometer (Shimadzu UV-2600Document5 pagesSOP UV-VIS Spectrophotometer (Shimadzu UV-2600valsquare valsquareNo ratings yet
- MOE Homlogy Modelling, Building and Evaluating The Protein StructureDocument13 pagesMOE Homlogy Modelling, Building and Evaluating The Protein Structureokta nursantiNo ratings yet
- Unsupervised Classfication Using ER MapperDocument9 pagesUnsupervised Classfication Using ER MapperavisenicNo ratings yet
- Guide to Radar 10.5 and EH 2.5 FeaturesDocument9 pagesGuide to Radar 10.5 and EH 2.5 FeaturesCelia SteimanNo ratings yet
- Spss Crosstabs Instructions Final DraftDocument9 pagesSpss Crosstabs Instructions Final Draftapi-669698004No ratings yet
- ASPEN TutorialDocument33 pagesASPEN TutorialAnoy Das MahapatraNo ratings yet
- Sqltools Tutorial enDocument12 pagesSqltools Tutorial ensasa68No ratings yet
- Diptrace TutorialDocument138 pagesDiptrace TutorialJose GutiNo ratings yet
- Ontology BuildingDocument7 pagesOntology BuildingEm Em,No ratings yet
- X'Pert HighScore Plus IntroductionDocument15 pagesX'Pert HighScore Plus IntroductionLevent Kartal100% (1)
- Tutorial Docking With GlideDocument5 pagesTutorial Docking With GlideJohn M. E. PurbaNo ratings yet
- QPA TrainingDocument13 pagesQPA TrainingsenthilkumarNo ratings yet
- ARCH348 Homework Help 5Document3 pagesARCH348 Homework Help 5yooga palanisamyNo ratings yet
- BLAST Algorithm Practical Assignment Detect Frameshift Mutations Structurally Similar ProteinsDocument5 pagesBLAST Algorithm Practical Assignment Detect Frameshift Mutations Structurally Similar Proteinslucy kabazziNo ratings yet
- Pymol Commands GatesDocument8 pagesPymol Commands GatesSandeep KaushikNo ratings yet
- L10 NCBI ExercisesDocument44 pagesL10 NCBI Exercisessrnagmote281980No ratings yet
- Pymol TutorialDocument21 pagesPymol TutorialpravindshawNo ratings yet
- An Introduction To NCBI BLAST: Prerequisites ResourcesDocument23 pagesAn Introduction To NCBI BLAST: Prerequisites ResourcesS ARUNANo ratings yet
- Excel 2007 for Scientists and EngineersFrom EverandExcel 2007 for Scientists and EngineersRating: 4 out of 5 stars4/5 (2)
- Excel for Scientists and EngineersFrom EverandExcel for Scientists and EngineersRating: 3.5 out of 5 stars3.5/5 (2)
- Journal of Molecular Structure: Mohammad Dinari, Fateme Gharahi, Parvin AsadiDocument8 pagesJournal of Molecular Structure: Mohammad Dinari, Fateme Gharahi, Parvin AsadiKarthi ShanmugamNo ratings yet
- 1 RK Assignment1Document1 page1 RK Assignment1Karthi ShanmugamNo ratings yet
- Om Sri Swamiye Sharanam AyyappaDocument4 pagesOm Sri Swamiye Sharanam AyyappaKarthi Shanmugam75% (4)
- Drug Discovery Historical Prescpective PDFDocument5 pagesDrug Discovery Historical Prescpective PDFKarthi ShanmugamNo ratings yet
- Tuesday, September 01, 2009 12:18 PM: Experimentation Page 1Document14 pagesTuesday, September 01, 2009 12:18 PM: Experimentation Page 1Karthi ShanmugamNo ratings yet
- 16-Week Module TemplateDocument53 pages16-Week Module TemplateKarthi ShanmugamNo ratings yet
- 1 RK Assignment1Document1 page1 RK Assignment1Karthi ShanmugamNo ratings yet
- Course: Medicinal Chemistry Course Code: BBICBI404R04/BICBI404R01 Instructor: Karthi ShanmugamDocument8 pagesCourse: Medicinal Chemistry Course Code: BBICBI404R04/BICBI404R01 Instructor: Karthi ShanmugamKarthi ShanmugamNo ratings yet
- TIMETABLE-Section A I II III IV V VI VII Viii IX X XI XII: PROGRAM: B.Tech.-Bioinformatics Semester: 4Document1 pageTIMETABLE-Section A I II III IV V VI VII Viii IX X XI XII: PROGRAM: B.Tech.-Bioinformatics Semester: 4Karthi ShanmugamNo ratings yet
- Interactive Syllabus Start1Document3 pagesInteractive Syllabus Start1Karthi ShanmugamNo ratings yet
- Research MethodsDocument11 pagesResearch MethodsKarthi ShanmugamNo ratings yet
- ReadingDocument5 pagesReadingKarthi ShanmugamNo ratings yet
- Strategies microbes use to invade the human bodyDocument6 pagesStrategies microbes use to invade the human bodyKarthi ShanmugamNo ratings yet
- F. Sanger, S. Nicklen and A.R. Coulson - DNA Sequencing With Chain-Terminating InhibitorsDocument5 pagesF. Sanger, S. Nicklen and A.R. Coulson - DNA Sequencing With Chain-Terminating InhibitorsYopghm698No ratings yet
- 1 RK Assignment1Document1 page1 RK Assignment1Karthi ShanmugamNo ratings yet
- MSC Unit 1Document16 pagesMSC Unit 1Karthi ShanmugamNo ratings yet
- Rasmol Quick Reference: Atom Expressions Display CommandsDocument2 pagesRasmol Quick Reference: Atom Expressions Display CommandsKarthi ShanmugamNo ratings yet
- Amino Ácid OsDocument2 pagesAmino Ácid OsSelene JuarezNo ratings yet
- Daily Ayyappan PoojaDocument12 pagesDaily Ayyappan Poojaperi9274No ratings yet
- Molecular Mechanics Force FieldsDocument26 pagesMolecular Mechanics Force FieldsKarthi ShanmugamNo ratings yet
- 30 FTPDocument6 pages30 FTPKarthiNo ratings yet
- Clustal W Reading PDFDocument2 pagesClustal W Reading PDFKarthi ShanmugamNo ratings yet
- ch08 JmolDocument12 pagesch08 JmolKarthi ShanmugamNo ratings yet
- MSC Unit 2Document9 pagesMSC Unit 2Karthi ShanmugamNo ratings yet
- Unit 5 Theory BiostDocument9 pagesUnit 5 Theory BiostKarthi ShanmugamNo ratings yet
- PymolDocument2 pagesPymolKarthi ShanmugamNo ratings yet
- Manual TR800Document40 pagesManual TR800Karthi ShanmugamNo ratings yet
- Nokia E6-00 UG enDocument132 pagesNokia E6-00 UG enKarthi ShanmugamNo ratings yet
- Fertigation For Vegetables: A Practical Guide For Small FieldsDocument7 pagesFertigation For Vegetables: A Practical Guide For Small FieldsSophia DwiratnaNo ratings yet
- Nivell C1 Anglés / Nivel C1 Inglés Comprensió Oral / Comprensión OralDocument8 pagesNivell C1 Anglés / Nivel C1 Inglés Comprensió Oral / Comprensión OralHiNo ratings yet
- Tissue processing problems and artifactsDocument18 pagesTissue processing problems and artifactsOsama Bakheet100% (2)
- Thermochemistry Heteroatomic CompoundsDocument7 pagesThermochemistry Heteroatomic CompoundsAnisatya AmaliahNo ratings yet
- The Story of Zilphy Claud and My Family Tree.Document36 pagesThe Story of Zilphy Claud and My Family Tree.Timothy100% (3)
- AlbuminaDocument4 pagesAlbuminaSuprovet LabotatorioNo ratings yet
- Haematology and TransfusionDocument3 pagesHaematology and TransfusionDinusha RajapakseNo ratings yet
- Prilabsa 2nd Workshop - MachalaDocument67 pagesPrilabsa 2nd Workshop - MachalaJunior Esquivel TorresNo ratings yet
- EnzymeDocument15 pagesEnzymeAljon Lara ArticuloNo ratings yet
- CR (Acac) ExptDocument4 pagesCR (Acac) Exptdsw27No ratings yet
- Queen of Philippine Orchids - The Rare and Beautiful Waling-waling FlowerDocument4 pagesQueen of Philippine Orchids - The Rare and Beautiful Waling-waling FlowerWendy BalaodNo ratings yet
- Anti AIDS HerbsDocument6 pagesAnti AIDS Herbskhansa maryamNo ratings yet
- Application Summary Sheet For Programme-Specific Requirements and Selection: Master's Programme in BiopharmaceuticalsDocument4 pagesApplication Summary Sheet For Programme-Specific Requirements and Selection: Master's Programme in BiopharmaceuticalsZikoraNo ratings yet
- Best Ayurvedic Medicine For Diabetes - Comparison & ReviewsDocument13 pagesBest Ayurvedic Medicine For Diabetes - Comparison & ReviewsSomanshu BanerjeeNo ratings yet
- Acceptability: ©WHO/Sergey VolkovDocument26 pagesAcceptability: ©WHO/Sergey VolkovBeatriz PatricioNo ratings yet
- Phasing With MRDocument95 pagesPhasing With MRLívia DantasNo ratings yet
- EYE Notes IgcseDocument3 pagesEYE Notes IgcseTay HermyNo ratings yet
- Mendel and The Principles of HeredityDocument61 pagesMendel and The Principles of Heredityfae-ar_raziNo ratings yet
- Hardy-Weinberg Principle WebquestDocument6 pagesHardy-Weinberg Principle Webquestapi-232072092No ratings yet
- CV Sample - PHDDocument3 pagesCV Sample - PHDJuhi DeoraNo ratings yet
- Genetic FingerprintingDocument9 pagesGenetic FingerprintingSumathi SelvarajNo ratings yet
- After DefenseDocument86 pagesAfter DefenseTsega YeNo ratings yet
- Worksheet Week 5 and 6Document16 pagesWorksheet Week 5 and 6EuniceNo ratings yet
- Lab Report Enzyme LabDocument7 pagesLab Report Enzyme LabrualrightNo ratings yet
- BIOFOSUN®Operating Procedure of Susceptibility Test PanelDocument19 pagesBIOFOSUN®Operating Procedure of Susceptibility Test PanelquankddNo ratings yet
- Kar Gut Kar 2017Document9 pagesKar Gut Kar 2017Fake TomNo ratings yet
- Ch. 25 Body Defence MechanismsDocument34 pagesCh. 25 Body Defence Mechanisms吴昊No ratings yet
- Psychology in Your Life 2nd Edition Grison Test BankDocument35 pagesPsychology in Your Life 2nd Edition Grison Test Bankluciaclark1mh0f100% (21)
- Algal Biorefinery - An Integrated Approach For Sustainable Biodiesel ProductionDocument16 pagesAlgal Biorefinery - An Integrated Approach For Sustainable Biodiesel ProductionCarlos GamarraNo ratings yet
- 20 Amino Acids in Human ProteinDocument2 pages20 Amino Acids in Human Proteinpanomo nasabyNo ratings yet