Professional Documents
Culture Documents
RNA Processing
Uploaded by
saeed313bbtOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
RNA Processing
Uploaded by
saeed313bbtCopyright:
Available Formats
RNA Processing or post
transcriptional modifications
Primary transcripts is linear, RNA copy of a
transcriptional unit.
Primary transcripts of both prokaryotic and
eukaryotic tRNA and rRNA are modified by
cleavage of the original transcripts by
ribonucleases.
Prokaryotic mRNA is generally identical to
itsprimary transcript, whereas eukaryotic
mRNA is extensivey modified.
A. rRNA
r- RNA of both prokaryotic and eukaryotic cells
aresynthesized from long precursor molecule
called preribosomal RNA.
From single precursor molecule 28S, 18S and 5.8S
rRNA are produced in eukaryotes and 23S, 16S
and 5S rRNA are produced in prokaryotes.
The nucleases that cleave and trim these
precursors of rRNA and tRNA are highly precise.
Primary Transcript
B. tRNA
tRNA precursors are converted into mature
tRNAs by a series of alterations:
cleavage of a 5 leader sequence
splicing to remove an intron
replacement of the 3 -terminal UU by CCA and
modification of several bases
Transfer RNA Precursor Processing
The conversion of a yeast tRNA precursor into a mature tRNA
requires the removal of a 14-nucleotide intron (yellow), the cleavage of a
5 leader (green), and the removal of UU and
the attachment of CCA at the 3 end (red). In addition, several bases are
modified.
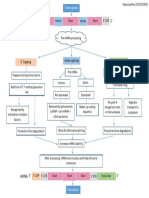
C. Eukaryotic mRNA
most extensively modified transcription
product is the product of RNA polymerase II:
The immediate product of an RNA polymerase
is sometimes referred to as pre-mRNA.
Most pre-mRNA molecules are spliced to
remove the introns.
both the 5 and the 3 ends are modified
1. 5 Capping
5 triphosphate end of the nascent RNA chain is
immediately modified.
First, a phosphate is released by hydrolysis.
The diphosphate 5 end then attacks the a-
phosphorus atom of GTP to form a very unusual 5
-5 triphosphate linkage. This distinctive terminus
is called a cap.
The N-7 nitrogen of the terminal guanine is then
methylated by S-adenosylmethionine.
Transfer RNA and ribosomal RNA molecules, in
contrast with messenger RNAs and small RNAs
that participate in splicing, do not have caps.
Caps contribute to the stability of mRNAs by
protecting their 5 ends from phosphatases
and nucleases.
In addition, caps enhance the translation of
mRNA by eukaryotic proteinsynthesizing
systems.
Capping the 5 End.
2. Poly-A tail
Pre-mRNA is also modified at the 3 end.
Most eukaryotic mRNAs contain a polyadenylate,
poly(A), tail at that end, added after transcription has
ended.
DNA does not encode this poly(A) tail.
Eukaryotic primary transcripts are cleaved by a specific
endonuclease that recognizes the sequence AAUAAA.
After cleavage by the endonuclease, a poly(A)
polymerase adds about 250 adenylate residues to the 3
end of the transcript; ATP is the donor in this reaction.
The role of the poly(A) tail is still not firmly established
despite much effort. However, evidence that it
enhances translation efficiency and the stability of
mRNA is accumulating.
Blocking the synthesis of the poly(A) tail by exposure to
3 -deoxyadenosine (cordycepin) does not interfere with
the synthesis of the primary transcript.
Messenger RNA devoid of a poly(A) tail can be
transported out of the nucleus. However, an mRNA
molecule devoid of a poly(A) tail is usually a much less
effective template for protein synthesis than is one
with a poly(A) tail.
Polyadenylation of a Primary Transcript
A specific endonuclease cleaves the RNA downstream of
AAUAAA. Poly(A) polymerase then adds about 250 adenylate residues
3. Removal of introns
Most genes in higher eukaryotes are composed
of exons and introns.
The introns must be excised and the exons linked
to form the final mRNA in a process called
splicing.
This splicing must be exquisitely sensitive: a one-
nucleotide slippage in a splice point would shift
the reading frame on the 3 side of the splice to
give an entirely different amino acid sequence.
Thus, the correct splice site must be clearly
marked.
The base sequences of thousands of intron- exon
junctions within RNA transcripts are known.
In eukaryotes from yeast to mammals, these
sequences have a common structural motif: the base
sequence of an intron begins with GU and ends with
AG.
The consensus sequence at the 5 splice in vertebrates
is AGGUAAGU.
At the 3 end of an intron, the consensus sequence is a
stretch of 10 pyrimidines (U or C), followed by any base
and then by C, and ending with the invariant AG.
Introns also have an important internal site
located between 20 and 50 nucleotides upstream
of the 3 splice site; it is called the branch site
In yeast, the branch site sequence is nearly
always UACUAAC, whereas in mammals a variety
of sequences are found.
The splicing of nascent mRNA molecules is a
complicated process.
It requires the cooperation of several small RNAs
and proteins that form a large complex called a
spliceosome
Splicing begins with the cleavage of the
phosphodiester bond between the upstream exon
(exon 1) and the 5 end of the Intron. The attacking
group in this reaction is the 2 -hydroxyl group of an
adenylate residue in the branch site. A 2 ,5 -
phosphodiester bond is formed between this A residue
and the 5 terminal phosphate of the intron. Hence a
branch is generated at this site, and a lariat
intermediate is formed.
The 3 -OH terminus of exon 1 then attacks the
phosphodiester bond between the intron and exon 2.
Exons 1 and 2 become joined, and the intron is
released in lariat form.
Splice Sites
Consensus sequences for the 5 splice site and the 3 splice site are shown.
Py stands for
pyrimidine.
Splicing Mechanism Used for mRNA Precursors.
Small Nuclear RNAs in Spliceosomes
Catalyze the Splicing of mRNA
Precursors
The nucleus contains many types of small RNA
molecules with fewer than 300 nucleotides, referred to
as snRNAs (small nuclear RNAs). A few of them
designated U1, U2, U4, U5, and U6 are essential for
splicing mRNA precursors. The
These RNA molecules are associated with specific
proteins to form complexes termed snRNPs (small
nuclear ribonucleoprotein particles); investigators often
speak of them as "snurps."
Spliceosomes are large (60S), dynamic assemblies
composed of snRNPs, other proteins called splicing
factors, and the mRNA precursors being processed
Spliceosome Assembly
U1 (blue) binds the 5 splice site and U2 (red) to the branch point. A preformed
U4-U5-U6 complex then joins the assembly to form the complete spliceosome
Alternative splicing
It is a widespread mechanism for generating
protein diversity.
The differential inclusion of exons into a mature
RNA, alternative splicing may be regulated to
produce distinct forms of a protein for specific
tissues or developmental stages.
recent estimates suggest that the RNA products
of 30% of human genes are alternatively spliced.
Alternative Splicing Pattern
A pre-mRNA with multiple exons is sometimes spliced in different ways.
Here, with two alternative exons (exons 2A and 2B) present, the mRNA can be
produced with neither, either, or both
exons included. More complex alternative splicing patterns also are possible
You might also like
- Genome Organization 1Document42 pagesGenome Organization 1nsjunnarkar100% (1)
- Solvent Miscibility and Polarity ChartDocument1 pageSolvent Miscibility and Polarity Chartdiptafara100% (2)
- Gene Silencing: An In-Depth Look at Mechanisms and ApplicationsDocument18 pagesGene Silencing: An In-Depth Look at Mechanisms and ApplicationsNAMRA RASHEEDNo ratings yet
- 18 DNA Structure and Replication-SDocument5 pages18 DNA Structure and Replication-Sjerhebeh100% (2)
- Transcription in ProkaryotesDocument16 pagesTranscription in ProkaryotesAditya Kanwal100% (1)
- Regulation of Gene Expression in Prokaryotic and Eukaryotic CellsDocument13 pagesRegulation of Gene Expression in Prokaryotic and Eukaryotic CellsMifta KhuljannahNo ratings yet
- DNA ReplicationDocument55 pagesDNA ReplicationZainab Jamal Siddiqui100% (1)
- DNA Footprinting: Pranjali Priya 15-MSVM 06 M.Sc. Biochemistry and Molecular BiologyDocument10 pagesDNA Footprinting: Pranjali Priya 15-MSVM 06 M.Sc. Biochemistry and Molecular BiologyPranjali Priya100% (1)
- General Translation MechanismDocument15 pagesGeneral Translation MechanismAishwarya KashyapNo ratings yet
- Transcription Translation Practice WorksheetDocument2 pagesTranscription Translation Practice WorksheetBesty Maranatha100% (1)
- Dna RepairDocument20 pagesDna RepairEaron Van JaboliNo ratings yet
- Replication, Transcription, Translation, Mutation HWDocument20 pagesReplication, Transcription, Translation, Mutation HWtahamidNo ratings yet
- Regulation of Gene Expression in Prokaryotes: © John Wiley & Sons, IncDocument65 pagesRegulation of Gene Expression in Prokaryotes: © John Wiley & Sons, IncRoberto CastroNo ratings yet
- DNA Manipulative EnzymesDocument17 pagesDNA Manipulative EnzymesZain Ul AbedienNo ratings yet
- Linkage: Harshraj Subhash Shinde KKW, Cabt, NashikDocument14 pagesLinkage: Harshraj Subhash Shinde KKW, Cabt, Nashiksivaram888No ratings yet
- Post Transcriptional ModificationDocument3 pagesPost Transcriptional ModificationLubainur RahmanNo ratings yet
- Regulation of Gene ExpressionDocument18 pagesRegulation of Gene ExpressionSH MannanNo ratings yet
- Transgenic AnimalsDocument13 pagesTransgenic AnimalsSamunderSinghSinghraNo ratings yet
- Genetics, Chapter 3, DNA Replication Lectures (Slides)Document115 pagesGenetics, Chapter 3, DNA Replication Lectures (Slides)Ali Al-QudsiNo ratings yet
- Post Transcriptional ModificationDocument31 pagesPost Transcriptional ModificationranasiddharthNo ratings yet
- TranscriptionDocument70 pagesTranscriptionkhan aishaNo ratings yet
- Biotecnology and Its ApplicationDocument87 pagesBiotecnology and Its ApplicationRangaswamyBiligiraiah100% (1)
- BY: Kanika Sabharwal Mtech BioinformaticsDocument21 pagesBY: Kanika Sabharwal Mtech BioinformaticsKanika SabharwalNo ratings yet
- RNA PolymeraseDocument14 pagesRNA PolymeraseKanaka lata SorenNo ratings yet
- Enhancer: Q1) What Is Role of Enhancers and Promoters in Transcription of Eukaryotes? AnsDocument9 pagesEnhancer: Q1) What Is Role of Enhancers and Promoters in Transcription of Eukaryotes? AnsSudeep BiswasNo ratings yet
- Structure and Function of DNA and RNADocument6 pagesStructure and Function of DNA and RNAEngr UmarNo ratings yet
- Post-Translational Modification: Dr. Md. Imtaiyaz HassanDocument61 pagesPost-Translational Modification: Dr. Md. Imtaiyaz Hassanbiotechnology2007100% (1)
- Gene Silencing: Presented by Aastha Pal M.Sc. 4 Semester (Biotechnology) Swami Rama Himalayan UniversityDocument22 pagesGene Silencing: Presented by Aastha Pal M.Sc. 4 Semester (Biotechnology) Swami Rama Himalayan UniversityAmit NegiNo ratings yet
- Western Blotting MicrsoftDocument45 pagesWestern Blotting MicrsoftSoNu de Bond100% (2)
- ReplicationDocument45 pagesReplicationAleena MustafaNo ratings yet
- Replication - Transcription - TranslationDocument75 pagesReplication - Transcription - TranslationJason FryNo ratings yet
- Gene Regulation: Mrs. Ofelia Solano SaludarDocument39 pagesGene Regulation: Mrs. Ofelia Solano SaludarmskikiNo ratings yet
- Purines, Pyrimidines and Nucleotides and the Chemistry of Nucleic AcidsFrom EverandPurines, Pyrimidines and Nucleotides and the Chemistry of Nucleic AcidsNo ratings yet
- Unit 7 Operon ConceptDocument18 pagesUnit 7 Operon ConceptSarah PavuNo ratings yet
- Nucleic AcidsDocument802 pagesNucleic AcidsTerrell Bowman100% (2)
- Lecture 33 Primer DesignDocument8 pagesLecture 33 Primer DesignJenna ScheiblerNo ratings yet
- PCR Types and Its ApplicationsDocument68 pagesPCR Types and Its ApplicationsShefali Pawar100% (1)
- Genomes in Prokaryotes and EukaryotesDocument46 pagesGenomes in Prokaryotes and EukaryoteskityamuwesiNo ratings yet
- Day 3 - Transcription and RNA ProcessingDocument50 pagesDay 3 - Transcription and RNA ProcessingAniket ParabNo ratings yet
- Transcription and RNA ProcessingDocument38 pagesTranscription and RNA ProcessingRishi Kumar100% (1)
- RNAi Gene Silencing for Plant Disease ManagementDocument43 pagesRNAi Gene Silencing for Plant Disease ManagementRuchi SharmaNo ratings yet
- Chapter#8 The Marketing Plan by Shepherd Hisrich, PetersDocument22 pagesChapter#8 The Marketing Plan by Shepherd Hisrich, Peterssaeed313bbt75% (4)
- Post Translational ModificationDocument4 pagesPost Translational ModificationSaira DogarNo ratings yet
- Rna Processing EukaryotesDocument33 pagesRna Processing EukaryotesNathanaelNo ratings yet
- Resna N K MicrobiologyDocument54 pagesResna N K MicrobiologyResna N K ResiNo ratings yet
- Gene Silencing - Antisense RNA & SiRNADocument18 pagesGene Silencing - Antisense RNA & SiRNAvenkatsuriyaprakash100% (1)
- Genetic RecombinationDocument60 pagesGenetic RecombinationJeevan BasnyatNo ratings yet
- DNA Replication in Prokaryotes and EukaryotesDocument7 pagesDNA Replication in Prokaryotes and Eukaryotessmn416No ratings yet
- Post Translational ModificationDocument11 pagesPost Translational ModificationCynCyntya HarlyanaNo ratings yet
- Dna Replication in EukaryotesDocument7 pagesDna Replication in EukaryotesAnish Shrestha100% (1)
- Prokaryotic Dna ReplicationDocument53 pagesProkaryotic Dna ReplicationRININo ratings yet
- L 10 Post Transcriptional ModificationDocument33 pagesL 10 Post Transcriptional ModificationsNo ratings yet
- Postranslational ModificationDocument78 pagesPostranslational ModificationnsjunnarkarNo ratings yet
- Linkage and crossing over: Genetic recombination between genes on homologous chromosomesDocument34 pagesLinkage and crossing over: Genetic recombination between genes on homologous chromosomesDhungana Surya RdNo ratings yet
- Process of Transcription in Eukaryotes ExplainedDocument1 pageProcess of Transcription in Eukaryotes ExplainedSomNo ratings yet
- Genetic Code HMDocument30 pagesGenetic Code HMdrhmpatel100% (2)
- Translational Control & Post-Translational ModificationsDocument10 pagesTranslational Control & Post-Translational ModificationsBhaskar GangulyNo ratings yet
- Post-Transcriptional Modification: An Overview of mRNA ProcessingDocument20 pagesPost-Transcriptional Modification: An Overview of mRNA ProcessingZain YaqoobNo ratings yet
- DNA Organization in Eukaryotic Chromosomes: Chapter 12: Section 12.4Document22 pagesDNA Organization in Eukaryotic Chromosomes: Chapter 12: Section 12.4Jaisy Samuel100% (1)
- Pre-mRNA: 5' Capping Polyadenylation Intron SplicingDocument1 pagePre-mRNA: 5' Capping Polyadenylation Intron SplicingKayoung HeoNo ratings yet
- Lecture 4 - Mechanism of Transcription in BacteriaDocument51 pagesLecture 4 - Mechanism of Transcription in BacteriaBakalJenazahNo ratings yet
- DNA Replication Prokaryotes EukaryotesDocument38 pagesDNA Replication Prokaryotes EukaryotesSudeep BiswasNo ratings yet
- GE IV TH SemDocument185 pagesGE IV TH Semrahul vivekNo ratings yet
- Gene Expression in ProkaryotesDocument43 pagesGene Expression in ProkaryotesravibiriNo ratings yet
- DNA Replication: How DNA Is CopiedDocument13 pagesDNA Replication: How DNA Is CopieddvdmegaNo ratings yet
- CHAPTER 12 Protein SynthesisDocument10 pagesCHAPTER 12 Protein SynthesisNadeem IqbalNo ratings yet
- RIBOZYMESDocument17 pagesRIBOZYMESSharan GayathrinathanNo ratings yet
- Lecture 2Document33 pagesLecture 2SUNDAS FATIMANo ratings yet
- Nutrient ReqDocument19 pagesNutrient Reqsaeed313bbtNo ratings yet
- AutophagyDocument7 pagesAutophagysaeed313bbtNo ratings yet
- Carbohydrates: Color Reactions and TestsDocument19 pagesCarbohydrates: Color Reactions and TestsAjith KumarNo ratings yet
- Global Financing Enterprise On Biotechnology.Document45 pagesGlobal Financing Enterprise On Biotechnology.saeed313bbtNo ratings yet
- Challenges in The Clinical Use of Genome Sequencing: Robert C. Green, Heidi L. Rehm, and Isaac S. KohaneDocument21 pagesChallenges in The Clinical Use of Genome Sequencing: Robert C. Green, Heidi L. Rehm, and Isaac S. Kohanesaeed313bbtNo ratings yet
- Nutrient ReqDocument19 pagesNutrient Reqsaeed313bbtNo ratings yet
- (Advances in Protein Chemistry 69) Wei Yang (Eds.) - DNA Repair and Replication-Elsevier, Academic Press (2004) PDFDocument396 pages(Advances in Protein Chemistry 69) Wei Yang (Eds.) - DNA Repair and Replication-Elsevier, Academic Press (2004) PDFSalma SalsabielaNo ratings yet
- Molecular Profiling of Some Barleria Species Using RBCL, Matk Gene Sequences and RAPD MarkersDocument4 pagesMolecular Profiling of Some Barleria Species Using RBCL, Matk Gene Sequences and RAPD MarkersShailendra RajanNo ratings yet
- Transcripción Alberts 2014 PDFDocument240 pagesTranscripción Alberts 2014 PDFLaliito LeNo ratings yet
- Rna Protein SynthesisDocument18 pagesRna Protein Synthesisapi-545652980No ratings yet
- Science-10 Q3 Lecture DNA and RNADocument2 pagesScience-10 Q3 Lecture DNA and RNAbruh669No ratings yet
- Genei™ Student RT PCR Teaching Kit ManualDocument11 pagesGenei™ Student RT PCR Teaching Kit ManualHemant KawalkarNo ratings yet
- DNA and RNA Are PolynucleotidesDocument35 pagesDNA and RNA Are PolynucleotidesnaomiNo ratings yet
- Reality of LifeDocument2 pagesReality of LifeDjela117No ratings yet
- Manatad - Exercise 3 - Nucleic Acids and ChromosomesDocument2 pagesManatad - Exercise 3 - Nucleic Acids and ChromosomesBryan Axel ManatadNo ratings yet
- Protein Synthesis ACEDocument14 pagesProtein Synthesis ACEZhiTing96No ratings yet
- PCR Master Mix ProtocolDocument2 pagesPCR Master Mix ProtocolDo Thi Van AnhNo ratings yet
- MAFFT Ver.7 - RBCL 2Document1 pageMAFFT Ver.7 - RBCL 2FJ-sonNo ratings yet
- Polymerase Chain ReactionDocument47 pagesPolymerase Chain ReactionshvnagaNo ratings yet
- DNA vs. RNA 5 Key Differences and ComparisonDocument6 pagesDNA vs. RNA 5 Key Differences and ComparisonRomalyn MoralesNo ratings yet
- Biology Unit 4 PDFDocument41 pagesBiology Unit 4 PDFsammam mahdi samiNo ratings yet
- Biochemistry, Polymerase Chain Reaction (PCR) - StatPearls - NCBI BookshelfDocument4 pagesBiochemistry, Polymerase Chain Reaction (PCR) - StatPearls - NCBI BookshelfsasaNo ratings yet
- 2 Replikasi DNADocument26 pages2 Replikasi DNAAngga WipraNo ratings yet
- MAN0012966 CloneJET PCR Cloning 40rxn UG PDFDocument2 pagesMAN0012966 CloneJET PCR Cloning 40rxn UG PDFPriya JainNo ratings yet
- DNA RepairDocument34 pagesDNA RepairAmit KaushikNo ratings yet
- Genetic Analysis An Integrated Approach 2Nd Edition Sanders Test Bank Full Chapter PDFDocument35 pagesGenetic Analysis An Integrated Approach 2Nd Edition Sanders Test Bank Full Chapter PDFallison.young656100% (14)
- Lecture Note #8 Genes & Genetic EngineeringDocument4 pagesLecture Note #8 Genes & Genetic Engineeringsev gsdNo ratings yet
- Protein Synthesis Home TaskDocument11 pagesProtein Synthesis Home TaskGraceEstoleCalo100% (1)
- Team Teaching Lesson PlanDocument2 pagesTeam Teaching Lesson Planapi-332987398No ratings yet
- Progress: Transcription by RNA Polymerase III: More Complex Than We ThoughtDocument5 pagesProgress: Transcription by RNA Polymerase III: More Complex Than We ThoughtDany Arrieta FloresNo ratings yet